8GDA
 
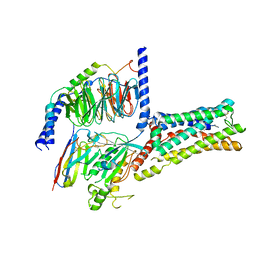 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (2S,5R)-5-[(1E,3S)-4,4-difluoro-3-hydroxy-4-phenylbut-1-en-1-yl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jiangqian, M, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EVQ
 
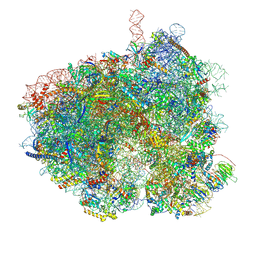 | | Hypopseudouridylated Ribosome bound with TSV IRES, eEF2, GDP, and sordarin, Structure I | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8G74
 
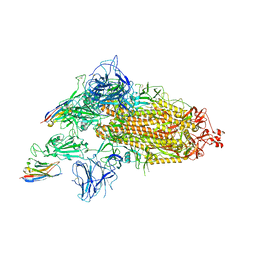 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
4XZ0
 
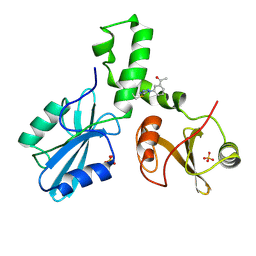 | | ZAP-70-tSH2:compound-A complex | | Descriptor: | 1-(3-{5-[(3-chlorobenzyl)sulfonyl]-1H-tetrazol-1-yl}phenyl)ethanone, SULFATE ION, Tyrosine-protein kinase ZAP-70 | | Authors: | Barros, T, Kuriyan, J, Visperas, P.R, Winger, J.A. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification by covalent reaction or oxidation of cysteine residues in the tandem-SH2 domains of ZAP-70 and Syk can block phosphopeptide binding.
Biochem. J., 465, 2015
|
|
8EVR
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure II | | Descriptor: | (1S,3S,3aR,4S,4aR,7R,7aR,8aS)-8a-{[(6-deoxy-4-O-methyl-alpha-D-altropyranosyl)oxy]methyl}-4-formyl-7-methyl-3-(propan-2-yl)decahydro-1,4-methano-s-indacene-3a(1H)-carboxylate, 18S rRNA, 25S rRNA, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8V1U
 
 | |
6ZHT
 
 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
7Q7S
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-1,3-dimethyl-2-oxidanylidene-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8B6H
 
 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B8H
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from M. tuberculosis in complex with DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, Alanine racemase, ... | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
4XU1
 
 | |
6ZRT
 
 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
7Q7T
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[(2S)-2,7-dimethyl-5,6-bis(oxidanylidene)-2,3-dihydro-1H-[1,4]oxazepino[6,5-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QZD
 
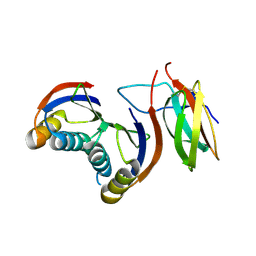 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikF with an engineered HMA domain of Pikp-1 (Pikp-SNK-EKE) from rice (Oryza sativa) | | Descriptor: | Avr-Pik, Resistance protein Pikp-1 | | Authors: | Maidment, J.H.R, Franceschetti, M, Longya, A, Banfield, M.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor.
Elife, 12, 2023
|
|
8HOM
 
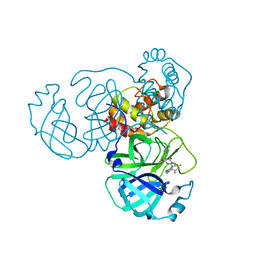 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
7QPX
 
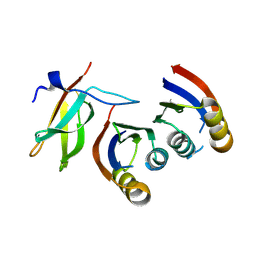 | |
7AMD
 
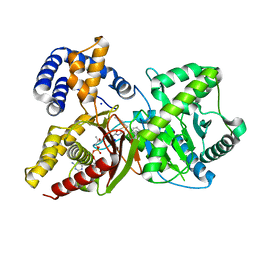 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
4Y4M
 
 | | Thiazole synthase Thi4 from Methanocaldococcus jannaschii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Putative ribose 1,5-bisphosphate isomerase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R)-2,3,5-tris(oxidanyl)-4-oxidanylidene-pentyl] hydrogen phosphate | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8UX6
 
 | | Structure of Fab201 with a T. parva sporozoite neutralizing B cell epitope of p67 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Gopalsamy, A, Fellouse, F.A, Miersch, S, Sidhu, S.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular characterization of a synthetic neutralizing antibody targeting p67 of Theileria parva.
Protein Sci., 34, 2025
|
|
6P9F
 
 | | Crystal structure of RAR-related orphan receptor C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 chimera, trans-4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)phenyl]pyrrolidine-1-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-06-10 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of potent, selective and orally bioavailable phenyl ((R)-3-phenylpyrrolidin-3-yl)sulfone analogues as ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7QPC
 
 | | Inward-facing NPA bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
7QLP
 
 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4Y4B
 
 | | Endothiapepsin in complex with fragment 323 | | Descriptor: | 6,6,8-trimethyl-5,6-dihydro[1,3]dioxolo[4,5-g]quinoline, ACETATE ION, Endothiapepsin, ... | | Authors: | Fu, K, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
