3WXO
 
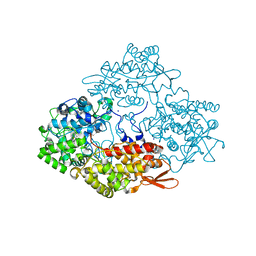 | | Crystal structure of isoniazid bound KatG catalase peroxidase from Synechococcus elongatus PCC7942 | | Descriptor: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Wada, K, Tada, T, Kamachi, S. | | Deposit date: | 2014-08-04 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of isoniazid-bound KatG catalase-peroxidase from Synechococcus elongatus PCC7942.
Febs J., 282, 2015
|
|
3B8X
 
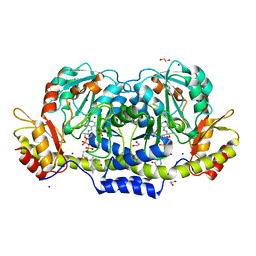 | | Crystal structure of GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) H188N mutant with bound GDP-perosamine | | Descriptor: | 1,2-ETHANEDIOL, Pyridoxamine 5-phosphate-dependent dehydrase, SODIUM ION, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GDP-4-Keto-6-deoxy-D-mannose 3-Dehydratase, Accommodating a Sugar Substrate in the Active Site.
J.Biol.Chem., 283, 2008
|
|
3RJE
 
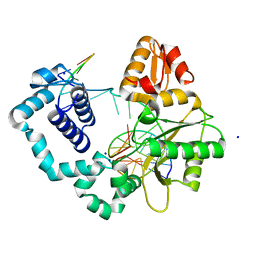 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RN0
 
 | |
3WSU
 
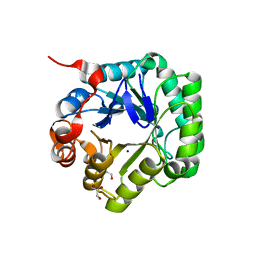 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
1XPE
 
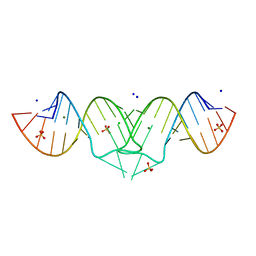 | | HIV-1 subtype B genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
3TJL
 
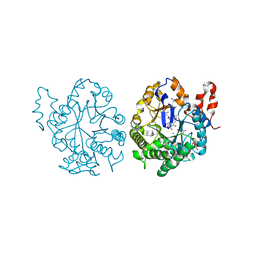 | |
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3ISC
 
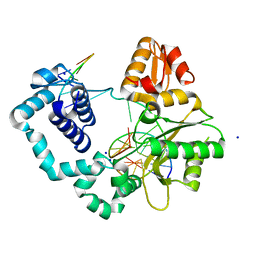 | | Binary complex of human DNA polymerase beta with an abasic site (THF) in the gapped DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(3DR)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
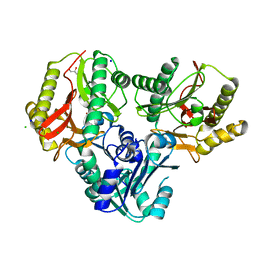
2ZSG
 
 | |
2ZNB
 
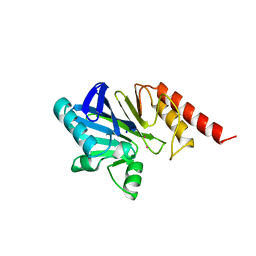 | | METALLO-BETA-LACTAMASE (CADMIUM-BOUND FORM) | | Descriptor: | CADMIUM ION, METALLO-BETA-LACTAMASE, SODIUM ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 6, 1997
|
|
3NRB
 
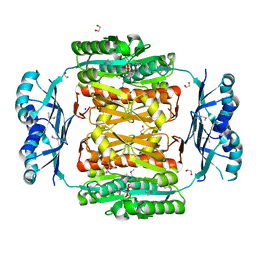 | |
1ZFA
 
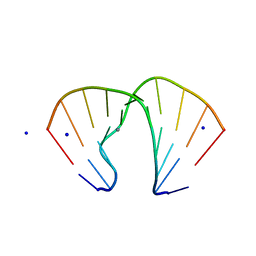 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UXK
 
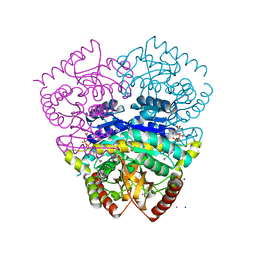 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
6SBL
 
 | | Human Carbonic Anhydrase II in complex with 4-hexylbenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-hexylbenzenesulfonamide, CITRATE ANION, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-07-21 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Human Carbonic Anhydrase II in complex with 4-hexylbenzenesulfonamide
To Be Published
|
|
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
6SBQ
 
 | | The crystal structure of PfA-M1 in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | DI(HYDROXYETHYL)ETHER, M1-family alanyl aminopeptidase, MALONATE ION, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
2FBB
 
 | | Crystal Structure Analysis of Hexagonal Lysozyme | | Descriptor: | Lysozyme C, NITRATE ION, SODIUM ION | | Authors: | Brinkmann, C, Weiss, M.S, Weckert, E. | | Deposit date: | 2005-12-09 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structure of the hexagonal crystal form of hen egg-white lysozyme.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5U8H
 
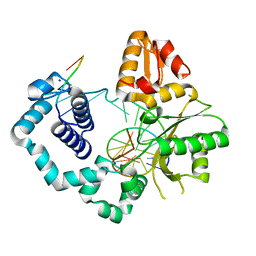 | | DNA Polymerase Beta G231D crystallized in PEG 400 | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Remote Mutations Induce Functional Changes in Active Site Residues of Human DNA Polymerase beta.
Biochemistry, 56, 2017
|
|
6SAC
 
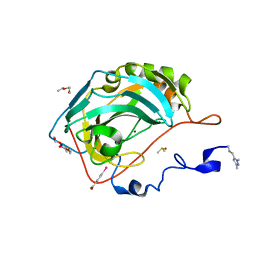 | | N-terminal expression tag remainder of human Carbonic Anhydrase II covalently modified by fragment | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 1-(1,3-dimethyl-1H-pyrazol-5-yl)methanamine, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
5UD1
 
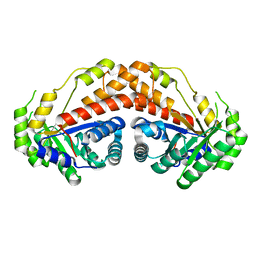 | |
5TWS
 
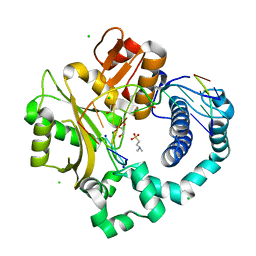 | | Post-catalytic complex of human Polymerase Mu (H329A) with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
6SYC
 
 | | Crystal structure of the lysozyme in presence of bromophenol blue at pH 6.5 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Lysozyme, ... | | Authors: | Camara-Artigas, A, Plaza-Garrido, M, Salinas-Garcia, M.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Lysozyme crystals dyed with bromophenol blue: where has the dye gone?
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5TXQ
 
 | |
