3DM5
 
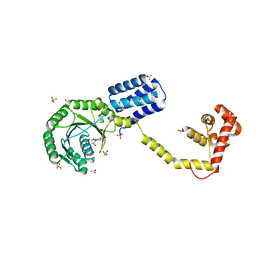 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DN4
 
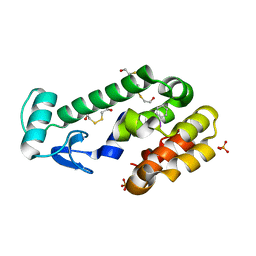 | |
3DQ9
 
 | |
3IFF
 
 | | 2'-SeMe-A modified DNA decamer | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*GP*CP*GP*TP*(XUA)P*C)-3'), MAGNESIUM ION | | Authors: | Sheng, J, Gan, J, Salon, J, Huang, Z. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selenium modification at 2'-position of adenosine for nucleic acid structure studies
To be Published
|
|
3DQM
 
 | |
2V3Y
 
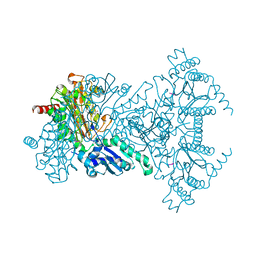 | |
3DRH
 
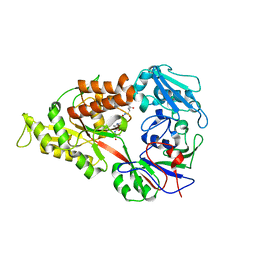 | | Crystal structure of Lactococcal OppA co-crystallized with Leu-enkephalin in an open conformation | | Descriptor: | Oligopeptide-binding protein oppA, peptide AAAAAA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
2VGL
 
 | | AP2 CLATHRIN ADAPTOR CORE | | Descriptor: | ADAPTOR PROTEIN COMPLEX AP-2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA-1, ... | | Authors: | Owen, D.J, Collins, B.M, McCoy, A.J, Evans, P.R. | | Deposit date: | 2007-11-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture and Functional Model of the Endocytic Ap2 Complex
Cell(Cambridge,Mass.), 109, 2002
|
|
3IHC
 
 | |
3DMO
 
 | |
2VJE
 
 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans.
Cell Death Differ., 15, 2008
|
|
2VH7
 
 | |
2VIO
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-3,5-dichlorobenzoic acid, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
3DQ2
 
 | |
3DQC
 
 | |
3DRF
 
 | | Lactococcal OppA complexed with an endogenous peptide in the closed conformation | | Descriptor: | Oligopeptide-binding protein oppA, endogenous peptide | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DVN
 
 | |
3DUF
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DZN
 
 | |
3DKR
 
 | |
3I8Z
 
 | | Crystal structure of human chromobox homolog 4 (CBX4) | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Amaya, M.F, Zhihong, L, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of human chromobox homolog 4 (CBX4)
To be Published
|
|
3DMD
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3DN0
 
 | | Pentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3E0A
 
 | |
3E0I
 
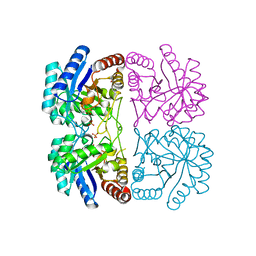 | | Cu2+ substituted Aquifex aeolicus KDO8PS in complex with PEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Gatti, D.L. | | Deposit date: | 2008-07-31 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
