8UGN
 
 | | In-situ structure of typeO supercomplex in respiratory chain (composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
3IJ6
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | Descriptor: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
8B8E
 
 | | Wild-type GH11 from Blastobotrys mokoenaii | | Descriptor: | 1,2-ETHANEDIOL, BmGH11, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coleman, T, Ravn, J.L, Larsbrink, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Yeasts Have Evolved Divergent Enzyme Strategies To Deconstruct and Metabolize Xylan.
Microbiol Spectr, 11, 2023
|
|
5IHN
 
 | |
8UGJ
 
 | | In-situ structure of typeB supercomplex in respiratory chain (composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
5IMZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F7 | | Descriptor: | Bacterioferritin comigratory protein, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IO2
 
 | | Xanthomonas campestris Peroxiredoxin Q - C48S mutant | | Descriptor: | Bacterioferritin comigratory protein, PHOSPHATE ION, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
3EOR
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, GERANYL DIPHOSPHATE, SODIUM ION, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Dawson, A. | | Deposit date: | 2008-09-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
5HSX
 
 | |
3E42
 
 | | Q138F HincII bound to GTCGAC and Ca2+ (cocrystallized) | | Descriptor: | 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Horton, N.C, Babic, A.C, Little, E.J, Manohar, V.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | DNA distortion and specificity in a sequence-specific endonuclease.
J.Mol.Biol., 383, 2008
|
|
8Y90
 
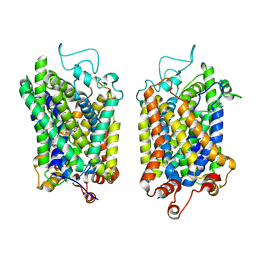 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
3USI
 
 | |
8Y93
 
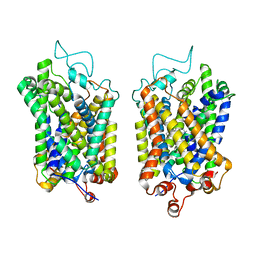 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8BAZ
 
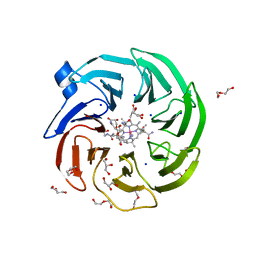 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittaker, J, Martinez-Felices, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobinamide.
To Be Published
|
|
8BB0
 
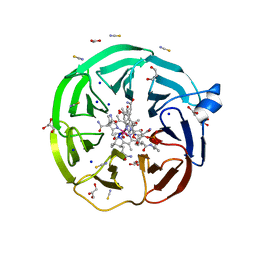 | | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobalamin. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J, Felices Martinez, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobinamide.
To Be Published
|
|
3ENZ
 
 | | Arsenolytic structure of Plasmodium falciparum purine nucleoside phosphorylase with hypoxanthine, ribose and arsenate ion | | Descriptor: | 1,4-anhydro-D-ribitol, ARSENATE, FORMIC ACID, ... | | Authors: | Chaikuad, A, Brady, R.L. | | Deposit date: | 2008-09-26 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
Bmc Struct.Biol., 9, 2009
|
|
8Y8Z
 
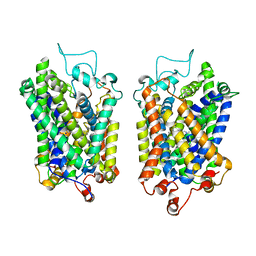 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y95
 
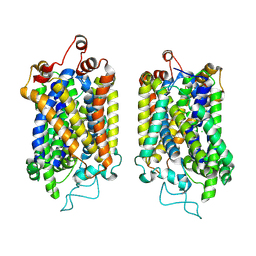 | | Structure of NET-NE in Occluded state | | Descriptor: | CHLORIDE ION, Noradrenaline, SODIUM ION, ... | | Authors: | Zhang, H, Xu, H.E, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
5I8U
 
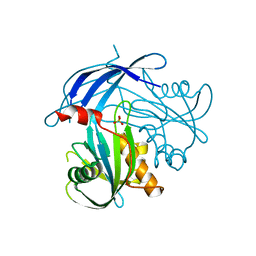 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
5I73
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central and allosteric sites | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
3EB5
 
 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
8B9Z
 
 | | Drosophila melanogaster complex I in the Active state (Dm1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Agip, A.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
5IDD
 
 | | Comment on S. W. M. Tanley and J. R. Helliwell Structural dynamics of cisplatin binding to histidine in a protein Struct. Dyn. 1, 034701 (2014) regarding the refinement of 4mwk, 4mwm, 4mwn and 4oxe and the method we have adopted. | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Helliwell, J.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn., 3, 2016
|
|
3MZG
 
 | | Crystal structure of a human prolactin receptor antagonist in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
8ZMU
 
 | |
