1NIM
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAK PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAN
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAO
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1KB7
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1NIL
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAK PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1KB8
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1TOX
 
 | | DIPHTHERIA TOXIN DIMER COMPLEXED WITH NAD | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1995-10-06 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide.
Biochemistry, 35, 1996
|
|
1NUE
 
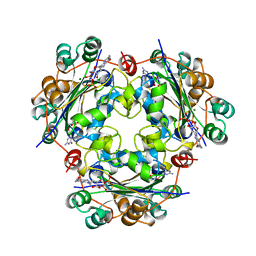 | | X-RAY STRUCTURE OF NM23 HUMAN NUCLEOSIDE DIPHOSPHATE KINASE B COMPLEXED WITH GDP AT 2 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Morera, S, Lacombe, M.-L, Yingwu, X, Lebras, G, Janin, J. | | Deposit date: | 1995-10-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution.
Structure, 3, 1995
|
|
1FFA
 
 | |
1FFC
 
 | |
1FFB
 
 | |
1FFE
 
 | |
1FFD
 
 | |
1LXA
 
 | | UDP N-ACETYLGLUCOSAMINE ACYLTRANSFERASE | | Descriptor: | UDP N-ACETYLGLUCOSAMINE O-ACYLTRANSFERASE | | Authors: | Roderick, S.L. | | Deposit date: | 1995-10-07 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase.
Science, 270, 1995
|
|
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
1LEY
 
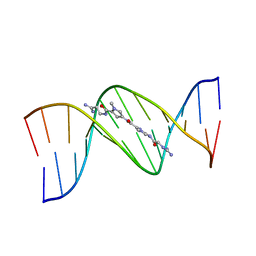 | | STRUCTURE OF A DICATIONIC MONOIMIDAZOLE LEXITROPSIN BOUND TO DNA (ORIENTATION 2) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MONOIMIDAZOLE LEXITROPSIN | | Authors: | Goodsell, D.S, Ng, H.L, Kopka, M.L, Lown, J.W, Dickerson, R.E. | | Deposit date: | 1995-10-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a dicationic monoimidazole lexitropsin bound to DNA.
Biochemistry, 34, 1995
|
|
1LEX
 
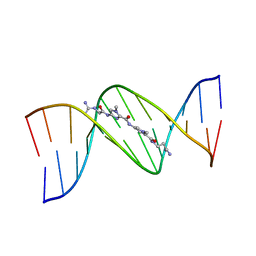 | | STRUCTURE OF A DICATIONIC MONOIMIDAZOLE LEXITROPSIN BOUND TO DNA (ORIENTATION 1) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MONOIMIDAZOLE LEXITROPSIN | | Authors: | Goodsell, D.S, Ng, H.L, Kopka, M.L, Lown, J.W, Dickerson, R.E. | | Deposit date: | 1995-10-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a dicationic monoimidazole lexitropsin bound to DNA.
Biochemistry, 34, 1995
|
|
1PIO
 
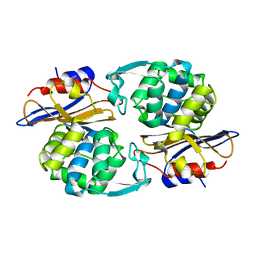 | |
1TOR
 
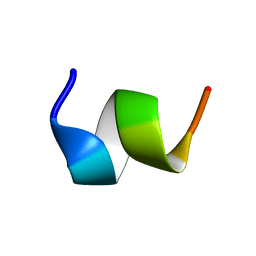 | | MOLECULAR DYNAMICS SIMULATION FROM 2D-NMR DATA OF THE FREE ACHR MIR DECAPEPTIDE AND THE ANTIBODY-BOUND [A76]MIR ANALOGUE | | Descriptor: | ACETYLCHOLINE RECEPTOR, MAIN IMMUNOGENIC REGION | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
2CSN
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH CKI7 | | Descriptor: | CASEIN KINASE-1, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, SULFATE ION | | Authors: | Xu, R.-M, Cheng, X. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for selectivity of the isoquinoline sulfonamide family of protein kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
