6B4M
 
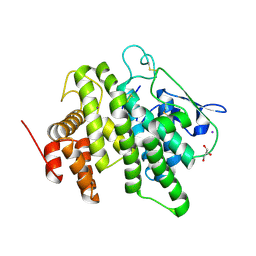 | | Structural characterization of a novel monotreme-specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Sharp, J.A, Kumar, A, Nicholas, K.R, Adams, T.E. | | Deposit date: | 2017-09-27 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1PQR
 
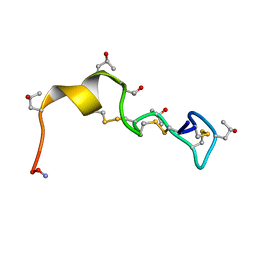 | | Solution Conformation of alphaA-Conotoxin EIVA | | Descriptor: | Alpha-A-conotoxin EIVA | | Authors: | Chi, S.-W, Park, K.-H, Suk, J.-E, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Conformation of alphaA-conotoxin EIVA, a Potent Neuromuscular Nicotinic Acetylcholine Receptor Antagonist from Conus ermineus
J.Biol.Chem., 278, 2003
|
|
8PF4
 
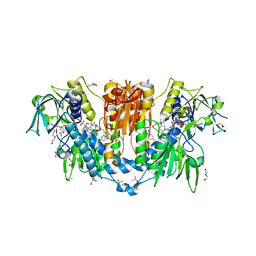 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)(4-fluorophenyl)carbamoyl)-1-methyl-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Exertier, C, Ilari, A, Fiorillo, A, Antonelli, L. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OEY
 
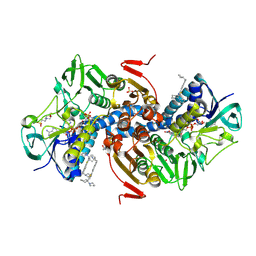 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-5-{5-[1-(Pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
1OMN
 
 | |
6XDK
 
 | |
6OEX
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor 3-(2-{1-[2-(Piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3- thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine | | Descriptor: | 3-(2-{1-[2-(piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
2P4L
 
 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2007-03-12 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
7N0Y
 
 | |
7N0W
 
 | |
7N43
 
 | |
7KNN
 
 | |
6PDK
 
 | |
6PPC
 
 | | Solution structure of conotoxin MiXXVIIA | | Descriptor: | Conopeptide phi-MiXXVIIA | | Authors: | Daly, N.L, Dekan, Z, Jin, A.H, Alewood, P.F. | | Deposit date: | 2019-07-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conotoxin phi-MiXXVIIA from the Superfamily G2 Employs a Novel Cysteine Framework that Mimics Granulin and Displays Anti-Apoptotic Activity.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6W7X
 
 | |
2JRY
 
 | | Structure and Sodium Channel Activity of an Excitatory I1-Superfamily Conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D, Babon, J, Yang, X, Fiedler, B, Chen, P, Yoshikami, D, Olivera, B, Bulaj, G, Norton, R. | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin.
Biochemistry, 46, 2007
|
|
6T98
 
 | | Trypanothione Reductase from Leishmania infantum in complex with 9a | | Descriptor: | 4-[3-methyl-1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-3-ium-4-yl]butan-1-amine, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
5XGL
 
 | | Co-crystal structure of Ac-AChBPP in complex with alpha-conotoxin LvIA | | Descriptor: | Alpha-conotoxin LvIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Xu, M.Y, Luo, S.L, Zhu, X.P. | | Deposit date: | 2017-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.439 Å) | | Cite: | The crystal structure of Ac-AChBP in complex with alpha-conotoxin LvIA reveals the mechanism of its selectivity towards different nAChR subtypes
Protein Cell, 8, 2017
|
|
4Y0I
 
 | | GABA-aminotransferase inactivated by conformationally-restricted inactivator | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Sanishvili, R, Lee, H.V, Dustin, D.H, Emma, D, Neil, K, Silverman, R.B, Liu, D. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | GABA-aminotransferase inactivated by conformationally-restricted inactivator
To Be Published
|
|
7S5E
 
 | |
8WKJ
 
 | |
6T95
 
 | | Trypanothione Reductase from Leismania infantum in complex with 4a | | Descriptor: | 1-[2-[5-[4-(4-azanylbutyl)-3-methyl-1,2,3-triazol-3-ium-1-yl]-2-[4-(2-phenylethyl)-1,3-thiazol-2-yl]phenoxy]ethyl]imidazolidin-2-one, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
