8J0O
 
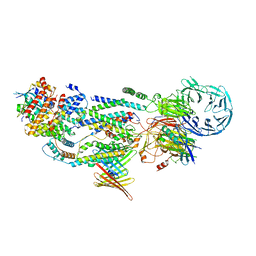 | | cryo-EM structure of human EMC and VDAC | | Descriptor: | ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ER membrane protein complex subunit 2, ... | | Authors: | Li, M, Zhang, C, Wu, J, Lei, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural insights into human EMC and its interaction with VDAC.
Aging (Albany NY), 16, 2024
|
|
6G6U
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 2.7 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, NITRATE ION, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
7QGJ
 
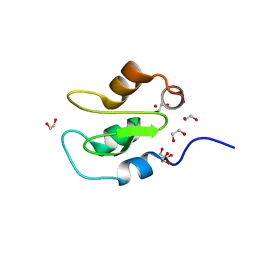 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
5K20
 
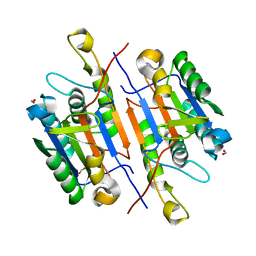 | | Caspase-7 S239E Phosphomimetic | | Descriptor: | Caspase-7 large subunit, Caspase-7 small subunit, FORMIC ACID | | Authors: | Eron, S.J, Hardy, J.A. | | Deposit date: | 2016-05-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PAK2 Phosphorylation Inhibits Caspase-7 by Two Divergent Mechanisms: Slowing Activation and Blocking Substrate Binding
To Be Published
|
|
1K86
 
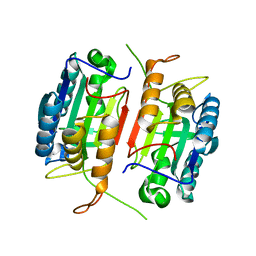 | | Crystal structure of caspase-7 | | Descriptor: | caspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1K88
 
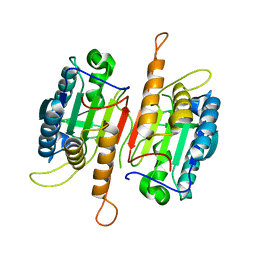 | | Crystal structure of procaspase-7 | | Descriptor: | procaspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
1U62
 
 | |
1VHN
 
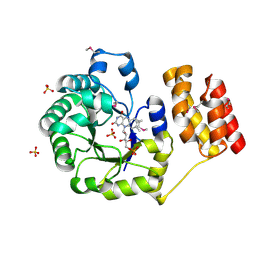 | |
6TIQ
 
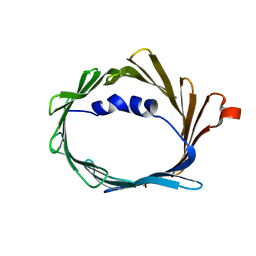 | |
1E2A
 
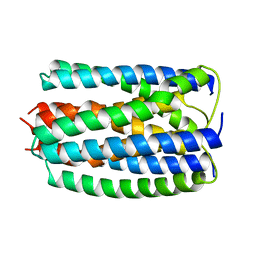 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
1EUD
 
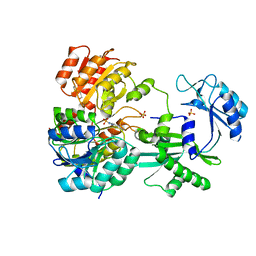 | | CRYSTAL STRUCTURE OF PHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
6OAJ
 
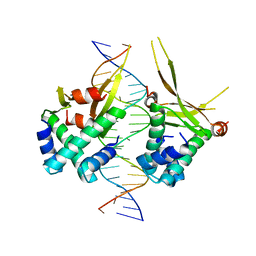 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
1EUC
 
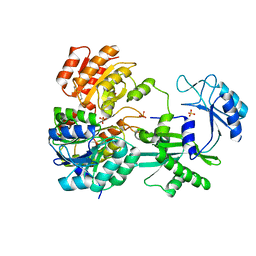 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | PHOSPHATE ION, SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
1F1J
 
 | |
6O8Q
 
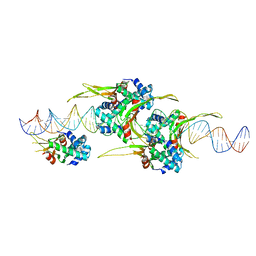 | | HUaa 19bp SYM DNA pH 4.5 | | Descriptor: | DNA (57-MER), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.216 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
6TIR
 
 | |
1KMC
 
 | |
6CL1
 
 | |
1LR1
 
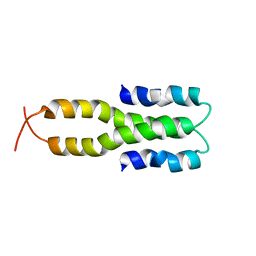 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
5CAE
 
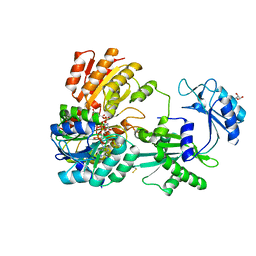 | |
1MUL
 
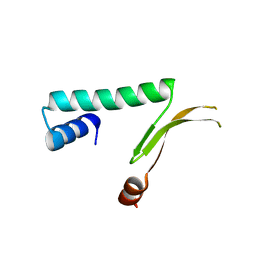 | | Crystal structure of the E. coli HU alpha2 protein | | Descriptor: | DNA binding protein HU-alpha | | Authors: | Ramstein, J, Hervouet, N, Coste, F, Zelwer, C, Oberto, J, Castaing, B. | | Deposit date: | 2002-09-24 | | Release date: | 2003-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence of a Thermal Unfolding Dimeric Intermediate for the Escherichia coli Histone-like HU Proteins: Thermodynamics and Structure.
J.Mol.Biol., 331, 2003
|
|
5DKO
 
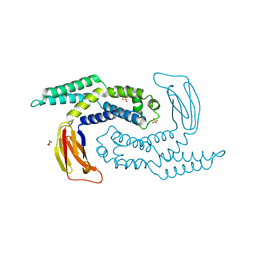 | | The structure of Escherichia coli ZapD | | Descriptor: | Cell division protein ZapD, SULFATE ION | | Authors: | Wroblewski, C, Kimber, M.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mutational Analyses of Escherichia coli ZapD Reveal Charged Residues Involved in FtsZ Filament Bundling.
J.Bacteriol., 198, 2016
|
|
1NI8
 
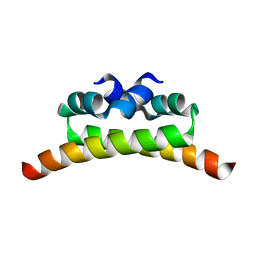 | | H-NS dimerization motif | | Descriptor: | DNA-binding protein H-NS | | Authors: | Bloch, V, Yang, Y, Margeat, E, Chavanieu, A, Aug, M.T, Robert, B, Arold, S, Rimsky, S, Kochoyan, M. | | Deposit date: | 2002-12-22 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The H-NS dimerisation domain defines a new fold contributing to DNA recognition
Nat.Struct.Biol., 10, 2003
|
|
1B2M
 
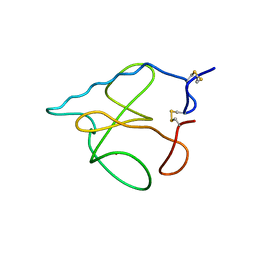 | | THREE-DIMENSIONAL STRUCTURE OF RIBONULCEASE T1 COMPLEXED WITH AN ISOSTERIC PHOSPHONATE ANALOGUE OF GPU: ALTERNATE SUBSTRATE BINDING MODES AND CATALYSIS. | | Descriptor: | 5'-R(*GP*(U34))-3', RIBONUCLEASE T1 | | Authors: | Arni, R.K, Watanabe, L, Ward, R.J, Kreitman, R.J, Kumar, K, Walz Jr, F.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with an isosteric phosphonate substrate analogue of GpU: alternate substrate binding modes and catalysis.
Biochemistry, 38, 1999
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
