4Q4L
 
 | |
6RD5
 
 | | CryoEM structure of Polytomella F-ATP synthase, focussed refinement of Fo and peripheral stalk, C2 symmetry | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
2RKW
 
 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | V-type ATP synthase beta chain | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
3P20
 
 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-10-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
3LG8
 
 | | Crystal structure of the C-terminal part of subunit E (E101-206) from Methanocaldococcus jannaschii of A1AO ATP synthase | | Descriptor: | A-type ATP synthase subunit E | | Authors: | Balakrishna, A.M, Manimekalai, M.S.S, Hunke, C, Gayen, S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal and solution structure of the C-terminal part of the Methanocaldococcus jannaschii A1AO ATP synthase subunit E revealed by X-ray diffraction and small-angle X-ray scattering
J.Bioenerg.Biomembr., 42, 2010
|
|
2KK7
 
 | |
3QG1
 
 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3SDZ
 
 | | Structural characterization of the subunit A mutant F427W of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2011-06-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Engineered tryptophan in the adenine-binding pocket of catalytic subunit A of A-ATP synthase demonstrates the importance of aromatic residues in adenine binding, forming a tool for steady-state and time-resolved fluorescence spectroscopy.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2D00
 
 | | Subunit F of V-type ATPase/synthase | | Descriptor: | CALCIUM ION, V-type ATP synthase subunit F | | Authors: | Makyio, H, Iino, R, Ikeda, C, Imamura, H, Tamakoshi, M, Iwata, M, Stock, D, Bernal, R.A, Carpenter, E.P, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2005-07-21 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
Embo J., 24, 2005
|
|
6TDW
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, peripheral stalk, rotational state 1 | | Descriptor: | ATPTB1, ATPTB3, ATPTB4, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
1MAB
 
 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
9JC2
 
 | | Engineering of ATP synthase Fo | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Hamaguchi-Suzuki, N, Ueno, H, Yasuda, K, Marui, R, Adachi, N, Senda, T, Noji, H, Murata, T. | | Deposit date: | 2024-08-28 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Engineering of ATP synthase for enhancement of proton-to-ATP ratio
Nat Commun, 2025
|
|
6PI4
 
 | |
5BQJ
 
 | |
6YNV
 
 | |
7YRY
 
 | | F1-ATPase of Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Grueber, G. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic insights of an up and down conformation of the Acinetobacter baumannii F 1 -ATPase subunit epsilon and deciphering the residues critical for ATP hydrolysis inhibition and ATP synthesis.
Faseb J., 37, 2023
|
|
5ZWL
 
 | |
8AP8
 
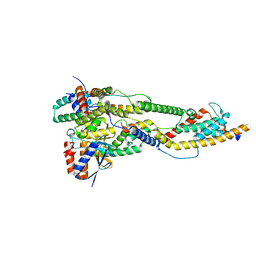 | | Peripheral stalk of Trypanosoma brucei mitochondrial ATP synthase | | Descriptor: | ATPTB3, ATPTB4, OSCP, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
6TDV
 
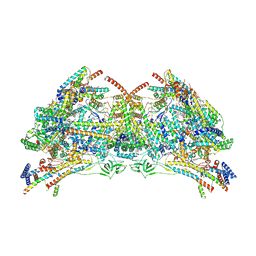 | |
9EVD
 
 | | In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Dietrich, L, Agip, A.N.A, Kuehlbrandt, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | In situ structure and rotary states of mitochondrial ATP synthase in whole Polytomella cells.
Science, 385, 2024
|
|
4XD7
 
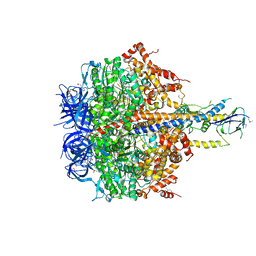 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
8ZI2
 
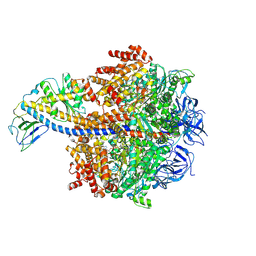 | | Cryo-EM reveals transition states of the Acinetobacter baumannii F1-ATPase rotary subunits gamma and epsilon and novel compound targets - Conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Le, K.C.M, Wong, C.F, Gruber, G. | | Deposit date: | 2024-05-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM reveals transition states of the Acinetobacter baumannii F 1 -ATPase rotary subunits gamma and epsilon , unveiling novel compound targets.
Faseb J., 38, 2024
|
|
