2G8U
 
 | |
4OZ0
 
 | |
2G92
 
 | |
6KLB
 
 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) | | Descriptor: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | Authors: | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7LVK
 
 | | Cfr-modified 50S subunit from Escherichia coli | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Stojkovic, V, Myasnikov, A.G, Frost, A, Fujimori, D.G. | | Deposit date: | 2021-02-25 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Investigating antibiotic resistance of a ribosomal-RNA methylating enzyme through directed evolution
To Be Published
|
|
6DCB
 
 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6DCC
 
 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
8EEX
 
 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
2G32
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8EMB
 
 | | X-ray crystal structure of Thermosynechococcus elongatus Si3 domain of RNA polymerase RpoC2 subunit | | Descriptor: | DNA-directed RNA polymerase subunit beta' | | Authors: | Murakami, K.S, Imashimizu, M, Qayyum, M.Z, Vishwakarma, R.K, Yuzenkova, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5E02
 
 | |
6Z6J
 
 | | Cryo-EM structure of yeast Lso2 bound to 80S ribosomes under native condition | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6K
 
 | | Cryo-EM structure of yeast reconstituted Lso2 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
2G1G
 
 | |
6DTI
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an unmodifed anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-06-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
8EA4
 
 | | V-K CAST Transpososome from Scytonema hofmanni, minor configuration | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, Cas12k, ... | | Authors: | Rizo, A.R, Park, J.-U, Tsai, A.W, Kellogg, E.K. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the holo CRISPR RNA-guided transposon integration complex.
Nature, 613, 2023
|
|
8EA3
 
 | | V-K CAST Transpososome from Scytonema hofmanni, major configuration | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, Cas12k, ... | | Authors: | Rizo, A.N, Park, J.-U, Tsai, A.W, Kellogg, E.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the holo CRISPR RNA-guided transposon integration complex.
Nature, 613, 2023
|
|
8S9T
 
 | | CRISPR-Cas type III-D effector complex | | Descriptor: | CRISPR RNA, Cas10, Cas7-2x, ... | | Authors: | Schwartz, E.A, Taylor, D.W. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Nat Commun, 15, 2024
|
|
4V7B
 
 | | Visualization of two tRNAs trapped in transit during EF-G-mediated translocation | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Lancaster, L, Sprink, T, Mielke, T, Loerke, J, Noller, H.F, Spahn, C.M.T. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6DB9
 
 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | Fab-Heavy-Chain, Fab-Light-Chain, MAGNESIUM ION, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6DB8
 
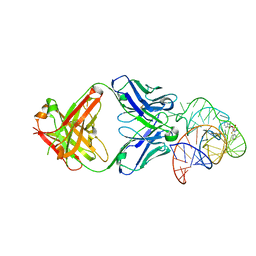 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | 2-[(Z)-(3-methyl-1,3-benzoxazol-2(3H)-ylidene)methyl]-3-(3-sulfopropyl)-1,3-benzothiazol-3-ium, Fab-Heavy chain, Fab-Light chain, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86541343 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
4V8E
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8D
 
 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8B
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-06 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
