1TCJ
 
 | |
1TCH
 
 | |
2C9T
 
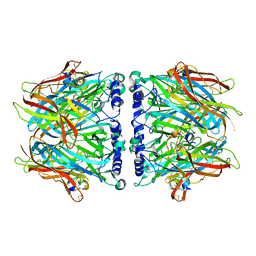 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CCO
 
 | |
2LXG
 
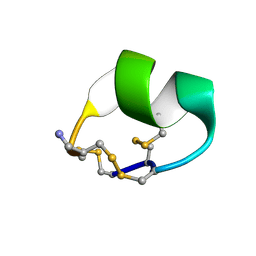 | |
2MTU
 
 | | Non-reducible analogues of alpha-conotoxin RgIA: [3,12]-trans dicarba RgIA | | Descriptor: | Dicarba Analogues of alpha-Conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S.D, Norton, R.S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
2MTT
 
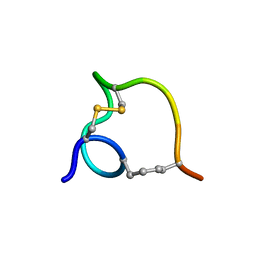 | | Non-reducible analogues of alpha-conotoxin RgIA: [3,12]-cis dicarba RgIA | | Descriptor: | Dicarba Analogues of alpha-Conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S.D, Norton, R.S. | | Deposit date: | 2014-08-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
4V3J
 
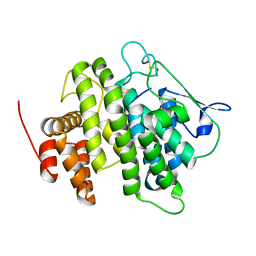 | | Structural and functional characterization of a novel monotreme- specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MONOTREME LACTATING PROTEIN | | Authors: | Kumar, A, Newman, J, Polekina, G, Adams, T.E, Sharp, J.A, Peat, T.S, Nicholas, K.R. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1P1P
 
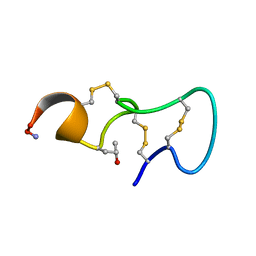 | | [PRO7,13] AA-CONOTOXIN PIVA, NMR, 12 STRUCTURES | | Descriptor: | AA-CONOTOXIN PIVA | | Authors: | Han, K.-H, Hwang, K.-J, Kim, S.-M, Kim, S.-K, Gray, W.R, Olivera, B.M, Rivier, J, Shon, K.J. | | Deposit date: | 1996-12-06 | | Release date: | 1997-07-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a novel conotoxin, [Pro 7,13] alpha A-conotoxin PIVA.
Biochemistry, 36, 1997
|
|
4V00
 
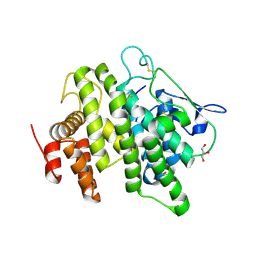 | | Structural and functional characterization of a novel monotreme- specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MONOTREME LACTATING PROTEIN | | Authors: | Enjapoori, A.K, Newman, J, Polekina, G, Adams, T.E, Sharp, J.A, Peat, T.S, Nicholas, K.R. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2LMZ
 
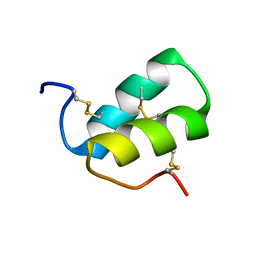 | |
2QBU
 
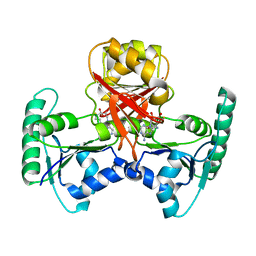 | | Crystal structure of Methanothermobacter thermautotrophicus CbiL | | Descriptor: | Precorrin-2 methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Frank, S, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidation of substrate specificity in the cobalamin (vitamin B12) biosynthetic methyltransferases. Structure and function of the C20 methyltransferase (CbiL) from Methanothermobacter thermautotrophicus.
J.Biol.Chem., 282, 2007
|
|
2FN6
 
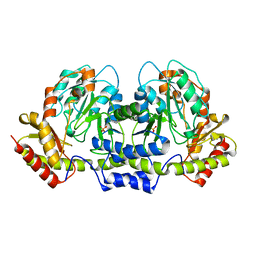 | | Helicobacter pylori PseC, aminotransferase involved in the biosynthesis of pseudoaminic acid | | Descriptor: | AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Cygler, M, Lunin, V.V, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2F8V
 
 | | Structure of full length telethonin in complex with the N-terminus of titin | | Descriptor: | N2B-Titin Isoform, SULFATE ION, Telethonin | | Authors: | Pinotsis, N, Petoukhov, M, Lange, S, Svergun, D, Zou, P, Gautel, M, Wilmanns, M. | | Deposit date: | 2005-12-04 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evidence for a dimeric assembly of two titin/telethonin complexes induced by the telethonin C-terminus.
J.Struct.Biol., 155, 2006
|
|
1ERB
 
 | |
1FEN
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | ALL-TRANS AXEROPHTHENE, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1HBP
 
 | |
1HBQ
 
 | |
1IIU
 
 | |
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1JLP
 
 | |
1KNL
 
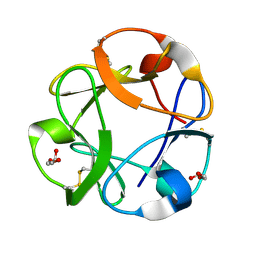 | | Streptomyces lividans Xylan Binding Domain cbm13 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1MC9
 
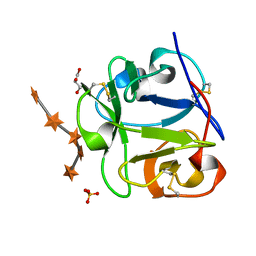 | | STREPROMYCES LIVIDANS XYLAN BINDING DOMAIN CBM13 IN COMPLEX WITH XYLOPENTAOSE | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, SULFATE ION, ... | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2002-08-06 | | Release date: | 2002-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KNM
 
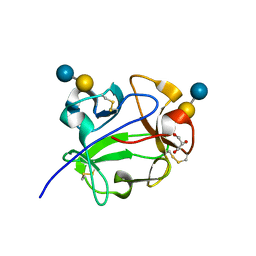 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
6CEG
 
 | |
