2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1KOC
 
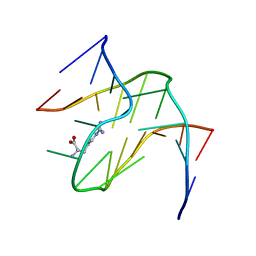 | | RNA APTAMER COMPLEXED WITH ARGININE, NMR | | Descriptor: | ARGININE, RNA (5'-R(P*AP*CP*AP*GP*GP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*CP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
9MMG
 
 | |
9MDX
 
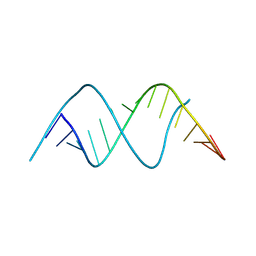 | |
9B9L
 
 | | RPRD1B C-terminal interacting domain bound to a pThr4 CTD peptide | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, SER-PRO-THR-SER-PRO-SER-TYR-SER-PRO-TPO-SER-PRO-SER-TYR-SER | | Authors: | Moreno, R.Y, Zhang, Y.J. | | Deposit date: | 2024-04-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thr 4 phosphorylation on RNA Pol II occurs at early transcription regulating 3'-end processing.
Sci Adv, 10, 2024
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
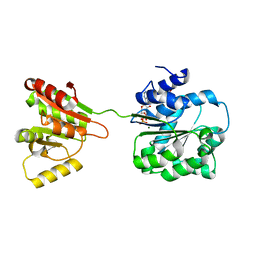 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
7WBX
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
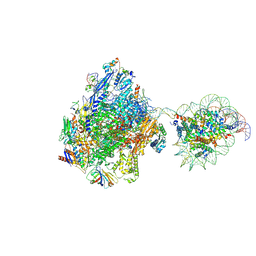 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8HE5
 
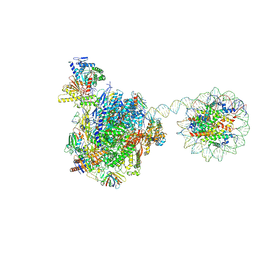 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBV
 
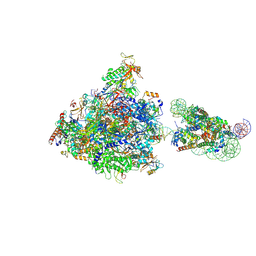 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | Descriptor: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
6INQ
 
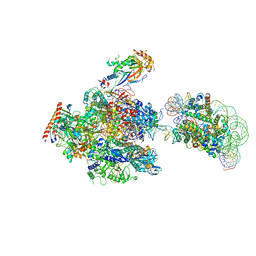 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5L
 
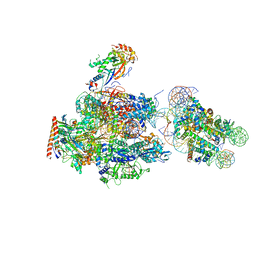 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5R
 
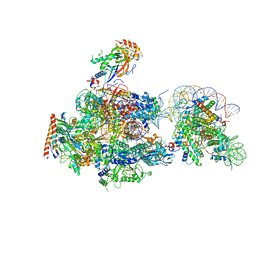 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5P
 
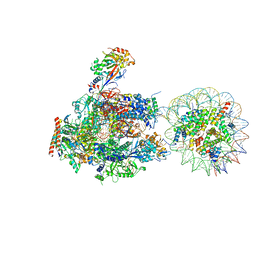 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5O
 
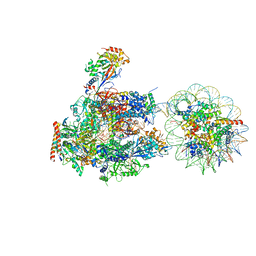 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5T
 
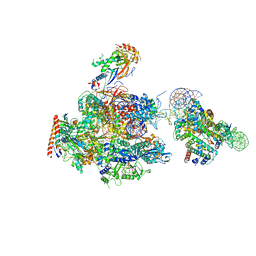 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5U
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
8HZM
 
 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
8HZE
 
 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
8HZF
 
 | | A new fluorescent RNA aptamer bound with N565 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-2-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-3-methyl-imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
8HZD
 
 | | A new fluorescent RNA aptamer bound with N618 | | Descriptor: | 4-[(~{E})-2-[(4~{Z})-4-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-1-methyl-5-oxidanylidene-imidazol-2-yl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, RNA (36-MER) | | Authors: | Song, Q.Q, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
8HZJ
 
 | | A new fluorescent RNA aptamer bound with N571 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
8HZK
 
 | | A new fluorescent RNA aptamer bound with N, iridium hexammine soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, DI(HYDROXYETHYL)ETHER, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 20, 2024
|
|
