6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
2C3K
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 4-[3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOL-6-YL]-2-METHOXYPHENOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2C3J
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | DEBROMOHYMENIALDISINE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFITE ION | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
6Z7F
 
 | |
6RQ3
 
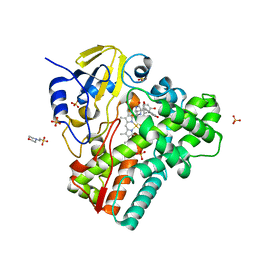 | | CYP121 in complex with 2,6-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(2,6-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6Z2V
 
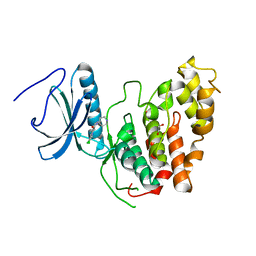 | | CLK3 A319V mutant bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6ZLN
 
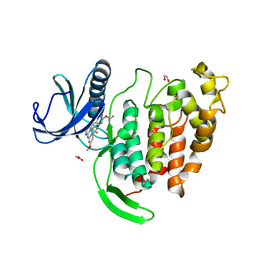 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6PYU
 
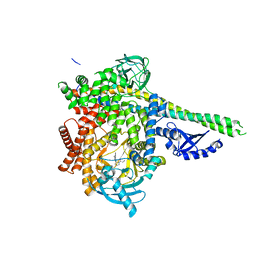 | | Human PI3Kdelta in complex with Compound 4-2 ((3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one) | | Descriptor: | (3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6ZB2
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK549 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZJF
 
 | | Crystal structure of STK17B (DRAK2) in complex with AP-229 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-cyclopropylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Picado, A, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
8E5U
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 9 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 4 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-nitro-4-(pyrimidin-2-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3P
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 5 | | Descriptor: | (2S,3S,4S,5R)-2-(hydroxymethyl)-1-{6-[3-nitro-5-(pyridin-4-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E4Z
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 8 | | Descriptor: | (2R,3R,4R,5S)-1-(6-{[(4P)-4-(5-cyclobutyl-1,2,4-oxadiazol-3-yl)-2-nitrophenyl]amino}hexyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ECW
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 11 | | Descriptor: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(pyridazin-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E4I
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 6 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{[(4P)-4-(5-methyl-1,2,4-oxadiazol-3-yl)-2-nitrophenyl]amino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E6G
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 10 | | Descriptor: | (2R,3R,4R,5S)-1-(6-{[(5M)-3-cyclopropyl-5-(pyridazin-3-yl)phenyl]amino}hexyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E4K
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 7 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[6-({(4M)-4-[2-(morpholin-4-yl)pyrimidin-4-yl]-2-nitrophenyl}amino)hexyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6TFP
 
 | | BTK in complex with LOU064, a potent and highly selective covalent inhibitor | | Descriptor: | SODIUM ION, Tyrosine-protein kinase BTK, ~{N}-[3-[6-azanyl-5-[2-[methyl(propanoyl)amino]ethoxy]pyrimidin-4-yl]-5-fluoranyl-2-methyl-phenyl]-4-cyclopropyl-2-fluoranyl-benzamide | | Authors: | Scheufler, C, Hinniger, A, Gutmann, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of LOU064 (Remibrutinib), a Potent and Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 63, 2020
|
|
6YYN
 
 | | Structure of Cathepsin S in complex with Compound 14 | | Descriptor: | CITRATE ANION, Cathepsin S, SULFATE ION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYO
 
 | | Structure of Cathepsin S in complex with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylsulfonylpiperazin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYP
 
 | | Structure of Cathepsin S in complex with Compound 2 | | Descriptor: | 1-(furan-2-ylmethyl)-5-(trifluoromethyl)benzimidazol-2-amine, ACETATE ION, Cathepsin S, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VJ3
 
 | | Carbonic Anhydrase II in complex with pyrimidine-based inhibitor | | Descriptor: | 2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2020-01-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells.
Eur.J.Med.Chem., 190, 2020
|
|
6YYQ
 
 | | Structure of Cathepsin S in complex with Compound 3 | | Descriptor: | (6~{R})-2-phenyl-5,6,7,8-tetrahydroquinazolin-6-amine, Cathepsin S | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYR
 
 | | Structure of Cathepsin S in complex with Compound 20b | | Descriptor: | (2~{R})-~{N}-(2-azanylideneethyl)-2-[2-(3-methyl-1,2-oxazol-5-yl)ethanoylamino]-3-(4-pyridin-2-ylpiperazin-1-yl)sulfonyl-propanamide, 1,2-ETHANEDIOL, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
