5WJS
 
 | |
8QAP
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with acetyl-pyrrole derivative compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(3,6,6-trimethyl-4-oxidanylidene-5,7-dihydro-2~{H}-isoindol-1-yl)-1,3-thiazol-2-yl]guanidine, Bromodomain-containing protein 4 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced cellular death in liver and breast cancer cells by dual BET/BRPF1 inhibitors.
Protein Sci., 33, 2024
|
|
8YP5
 
 | | The structure of MAP2K4 complexed with 5Z7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 4, MAGNESIUM ION | | Authors: | Yumura, S, Kinishita, T. | | Deposit date: | 2024-03-15 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conserved gatekeeper methionine regulates the binding and access of kinase inhibitors to ATP sites of MAP2K1, 4, and 7: Clues for developing selective inhibitors.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
6AC3
 
 | |
8QAL
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with acetyl-pyrrole derivative compound 83 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(3-methyl-4-oxidanylidene-2,5,6,7-tetrahydroisoindol-1-yl)-1,3-thiazol-2-yl]guanidine, Bromodomain-containing protein 4 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Enhanced cellular death in liver and breast cancer cells by dual BET/BRPF1 inhibitors.
Protein Sci., 33, 2024
|
|
6GZC
 
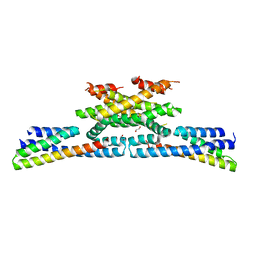 | | heterotetrameric katanin p60:p80 complex | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Katanin p60 ATPase-containing subunit A1, ... | | Authors: | Faltova, L, Jiang, K, Frey, D, Wu, Y, Capitani, G, Prota, A.E, Akhmanova, A, Steinmetz, M.O, Kammerer, R.A. | | Deposit date: | 2018-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Heterotetrameric Katanin p60:p80 Complex.
Structure, 27, 2019
|
|
8VHP
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to CDP and two molecules of ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
6PYA
 
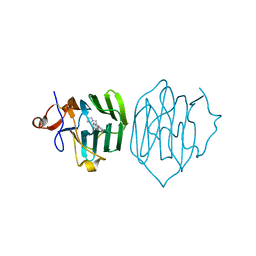 | |
8VTB
 
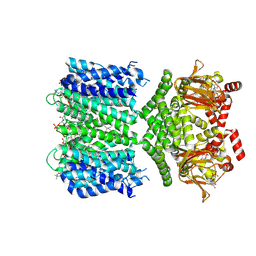 | | SthK R120A R124A in the presence of PIP2 and cAMP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, ... | | Authors: | Schmidpeter, P.A.M, Thon, O, Nimigean, C.M. | | Deposit date: | 2024-01-26 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | PIP2 inhibits pore opening of the cyclic nucleotide-gated channel SthK.
Nat Commun, 15, 2024
|
|
8QT2
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-6 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
7P41
 
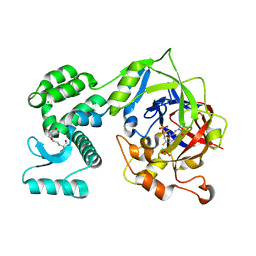 | | Crystal Structure of human mARC1 A165T Variant | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Mitochondrial amidoxime-reducing component 1,Endolysin,Mitochondrial amidoxime-reducing component 1, ... | | Authors: | Struwe, M.A, Scheidig, A.J. | | Deposit date: | 2021-07-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Letter to the editor: The clinically relevant MTARC1 p.Ala165Thr variant impacts neither the fold nor active site architecture of the human mARC1 protein.
Hepatol Commun, 6, 2022
|
|
6NBS
 
 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
8QS7
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 70 (1084352) | | Descriptor: | 1-[8-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
5MTJ
 
 | | Yes1-SH2 in complex with monobody Mb(Yes_1) | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Monobody Mb(Yes_1), SULFATE ION, ... | | Authors: | Sha, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
8QS8
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 78 (1084378) | | Descriptor: | 1-[8-(4-bromophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS9
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 83 (1084383) | | Descriptor: | 1-[8-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
8QSA
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 86 (1084384) | | Descriptor: | 1-[(5~{R})-2-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,7-diazaspiro[4.4]nonan-7-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8CMO
 
 | | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp. | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Fadeeva, M, Klaiman, D, Nelson, N. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp.
To Be Published
|
|
5X1K
 
 | | Vanillate/3-O-methylgallate O-demethylase, LigM, 3-O-methylgallate complex form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methoxy-4,5-bis(oxidanyl)benzoic acid, ... | | Authors: | Harada, A, Senda, T. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6
FEBS J., 284, 2017
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
6EPV
 
 | | The ATAD2 bromodomain in complex with compound 5 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-oxidanylidene-6,7-dihydro-5~{H}-1,3-benzothiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6Q5F
 
 | | OXA-48_P68A-CAZ. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.-K.S, Samuelsen, O. | | Deposit date: | 2018-12-08 | | Release date: | 2019-03-20 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
7QLQ
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NAD, AND DIMETHOXYPHENYL ACETAMIDE | | Descriptor: | 2-(3,4-dimethoxyphenyl)ethanamide, ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Sridhar, S, Kiema, T.R, Widersten, M, Wierenga, R.K. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
