2HMQ
 
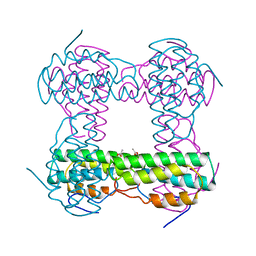 | |
2HMZ
 
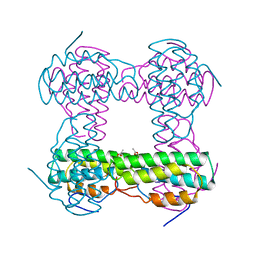 | |
1LB8
 
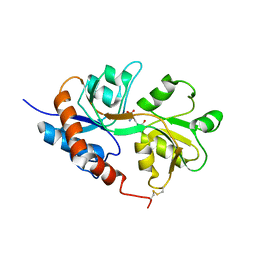 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1U65
 
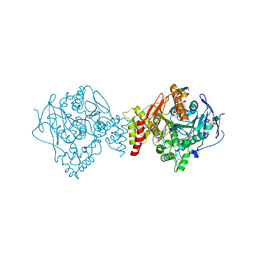 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
4URH
 
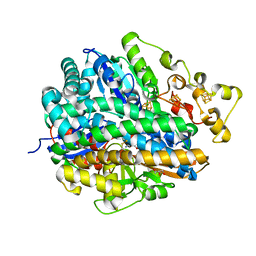 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
6IH1
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-GMP bound form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
5D1K
 
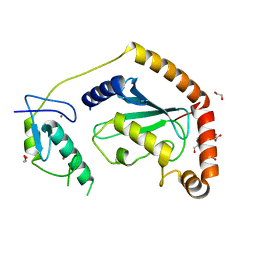 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
5CXX
 
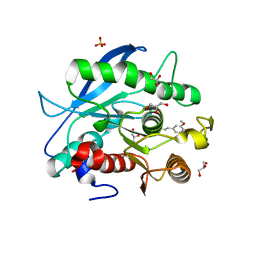 | | Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Gruninger, R.J, Abbott, D.W. | | Deposit date: | 2015-07-29 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Contributions of a unique beta-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases.
Biochem.J., 473, 2016
|
|
6IJ2
 
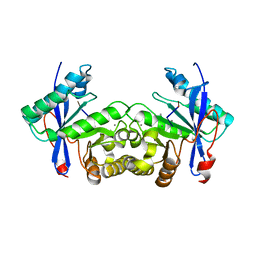 | |
8RQX
 
 | |
5NRI
 
 | |
9DZ8
 
 | | Catalytic domain of Dihydrolipoamide Succinytransferase | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | | Authors: | Carr, K.D, Borst, A.J, Weidle, C. | | Deposit date: | 2024-10-15 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Protein identification using Cryo-EM and artificial intelligence guides improved sample purification.
J Struct Biol X, 11, 2025
|
|
6IFQ
 
 | |
5NRH
 
 | |
4WB7
 
 | | Crystal structure of a chimeric fusion of human DnaJ (Hsp40) and cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WB8
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit), exon 1 deletion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4L8U
 
 | | X-ray study of human serum albumin complexed with 9 amino camptothecin | | Descriptor: | (2S)-2-[1-amino-8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]-2-hydroxybutanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-17 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
2HWT
 
 | |
2UZA
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Cavazza, C, Contreras-Martel, C, Fontecilla-Camps, J.C. | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate.
Structure, 14, 2006
|
|
3S43
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug amprenavir | | Descriptor: | GLYCEROL, IODIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Tie, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
9MRL
 
 | |
9MRN
 
 | |
9MRM
 
 | |
9MRK
 
 | |
7ST9
 
 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
