2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
5J2D
 
 | |
3NCE
 
 | | A mutant human Prolactin receptor antagonist H27A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
8P8G
 
 | | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Maslac, N, Wagner, T. | | Deposit date: | 2023-06-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity.
Jacs Au, 3, 2023
|
|
2OVL
 
 | |
8OVS
 
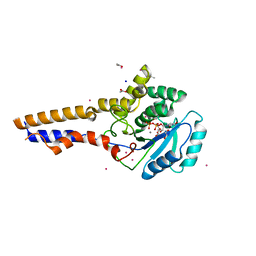 | |
8P3M
 
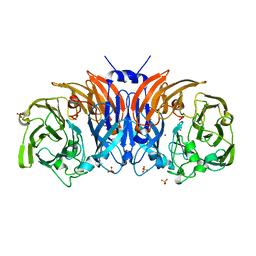 | | The structure of thiocyanate dehydrogenase mutant form with Lys 281 replaced by Ala from Thioalkalivibrio paradoxus | | Descriptor: | BORIC ACID, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Komolov, A.S, Rakitina, T.V, Dergousova, N.I, Dorovatovskii, P.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Improvement of the Diffraction Properties of Thiocyanate Dehydrogenase Crystals
Crystallography Reports, 2023
|
|
4E3Q
 
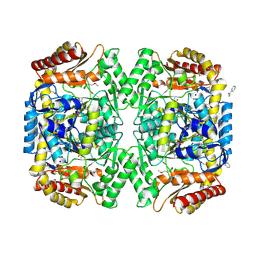 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
5IUB
 
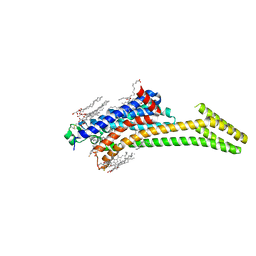 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12x at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
3DIY
 
 | |
4E6P
 
 | |
5I75
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central site and Br-citalopram at the allosteric site | | Descriptor: | (1S)-1-(4-bromophenyl)-1-[3-(dimethylamino)propyl]-1,3-dihydro-2-benzofuran-5-carbonitrile, (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
7A5G
 
 | | Structure of the elongating human mitoribosome bound to mtEF-Tu.GMPPCP and A/T mt-tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
8QRE
 
 | | Cholera holotoxin (wildtype) | | Descriptor: | ACETATE ION, Cholera enterotoxin subunit A, Cholera enterotoxin subunit B, ... | | Authors: | Mojica, N, Cordara, G, Krengel, U. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using Vibrio natriegens for High-Yield Production of Challenging Expression Targets and for Protein Perdeuteration.
Biochemistry, 63, 2024
|
|
7A6B
 
 | | 1.33 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.33 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
8PW6
 
 | | C respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
1P0Z
 
 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
5J2B
 
 | |
5J2H
 
 | |
7C1Z
 
 | | ATP bound structure of Pseudouridine kinase (PUKI) from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PfkB-like carbohydrate kinase family protein, ... | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09617043 Å) | | Cite: | Structural basis for the substrate specificity and catalytic features of pseudouridine kinase from Arabidopsis thaliana.
Nucleic Acids Res., 49, 2021
|
|
3EZQ
 
 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
2X8J
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | GLYCEROL, INTRACELLULAR SUBTILISIN PROTEASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Vedodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an intracellular subtilisin reveals novel structural features unique to this subtilisin family.
Structure, 18, 2010
|
|
5HK7
 
 | | Bacterial sodium channel pore, 2.95 Angstrom resolution | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, CHLORIDE ION, Ion transport protein, ... | | Authors: | Shaya, D, Findeisen, F, Rohaim, A, Minor, D.L. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
7A5F
 
 | | Structure of the stalled human mitoribosome with P- and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
8QRD
 
 | | OleP in complex with testosterone in high salt crystallization conditions | | Descriptor: | Cytochrome P-450, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with testosterone in high salt crystallization conditions
To Be Published
|
|
