8A1V
 
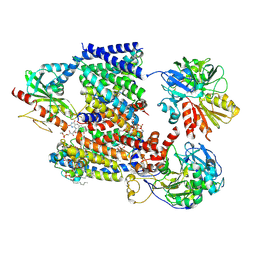 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1W
 
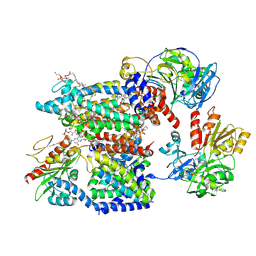 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1X
 
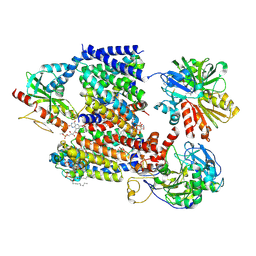 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1T
 
 | | Sodium pumping NADH-quinone oxidoreductase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
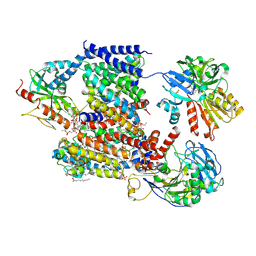 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6R7B
 
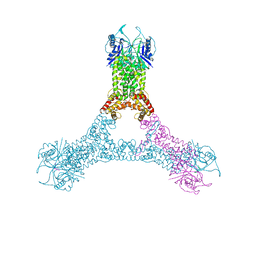 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6RA6
 
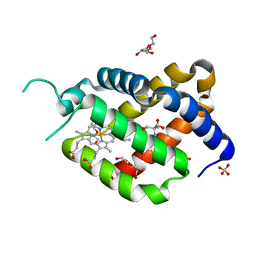 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
8A1U
 
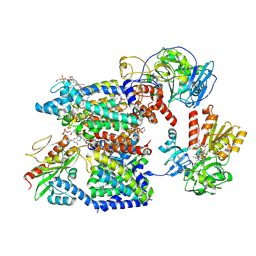 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1D1V
 
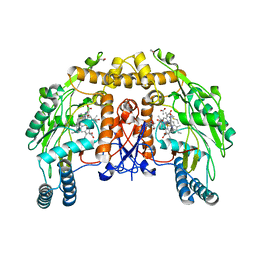 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH S-ETHYL-N-PHENYL-ISOTHIOUREA (H4B BOUND) | | Descriptor: | 2-ETHYL-1-PHENYL-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
8T6N
 
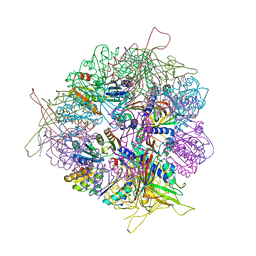 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8D44
 
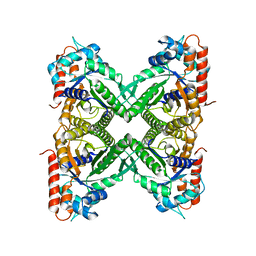 | |
6KXR
 
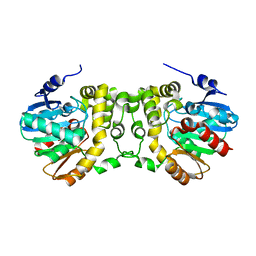 | | Crystal structure of wild type Alp1U from the biosynthesis of kinamycins | | Descriptor: | D-MALATE, Putative hydrolase | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45073676 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
1D62
 
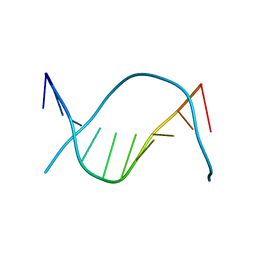 | |
7Q7Q
 
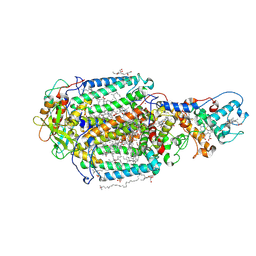 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Baath, P, Banacore, A, Neutze, R. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q7P
 
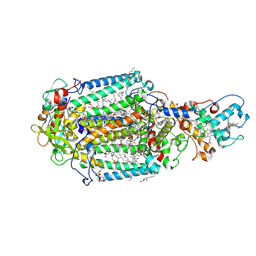 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Baath, P, Banacore, A, Neutze, R. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1LLA
 
 | |
8PAS
 
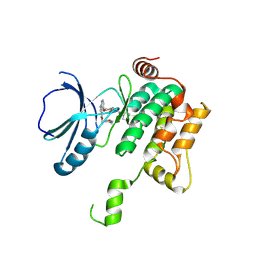 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | 4-[2,6-bis(fluoranyl)-4-(3-morpholin-4-ylpropylcarbamoylamino)phenoxy]-~{N}-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]-1~{H}-pyrrolo[2,3-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
6L7Q
 
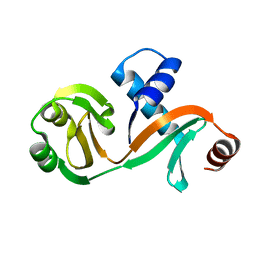 | |
5C4U
 
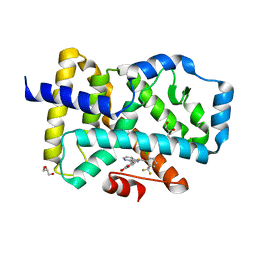 | |
1D61
 
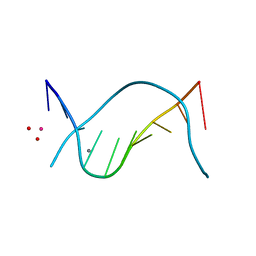 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | Descriptor: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
7CW0
 
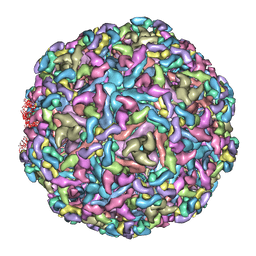 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5FCZ
 
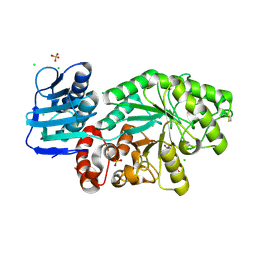 | |
6WMK
 
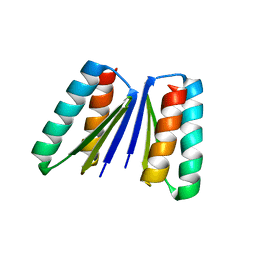 | | Crystal structure of beta sheet heterodimer LHD29 | | Descriptor: | Beta sheet heterodimer LHD29 - Chain A, Beta sheet heterodimer LHD29 - Chain B | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2020-04-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7CVZ
 
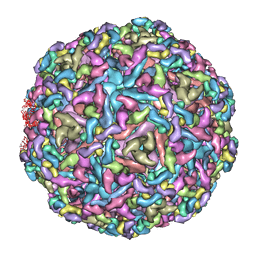 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CVY
 
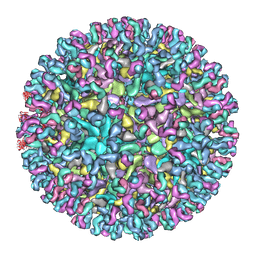 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-124 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
