7AM1
 
 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
6ZTU
 
 | |
2IL9
 
 | |
4F3M
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | 1,2-ETHANEDIOL, BH0337 protein, SULFATE ION | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System.
Structure, 20, 2012
|
|
2L9W
 
 | |
8QO4
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO2
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
1LC6
 
 | | Solution Structure of the U6 Intramolecular Stem-loop RNA | | Descriptor: | U6 Intramolecular Stem-loop RNA | | Authors: | Huppler, A, Nikstad, L.J, Allmann, A.M, Brow, D.A, Butcher, S.E. | | Deposit date: | 2002-04-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure.
Nat.Struct.Biol., 9, 2002
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
5UEG
 
 | | RNA primer-template complex with guanosine dinucleotide G(5')pppp(5')G ligand | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*GP*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3'), [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-01-02 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the mechanism of nonenzymatic RNA primer extension from the structure of an RNA-GpppG complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UEF
 
 | |
5UED
 
 | | RNA primer-template complex with guanosine dinucleotide ligand G(5')pp(5')G | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*GP*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3'), [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-01-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insight into the mechanism of nonenzymatic RNA primer extension from the structure of an RNA-GpppG complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WLH
 
 | |
3L26
 
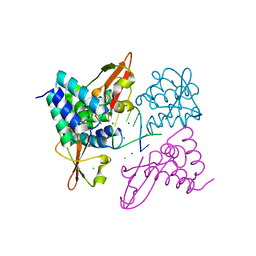 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4FF2
 
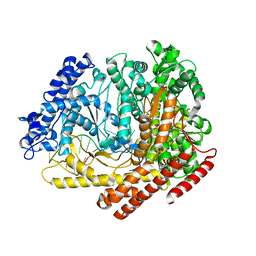 | | N4 mini-vRNAP transcription initiation complex, 2 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
3PRH
 
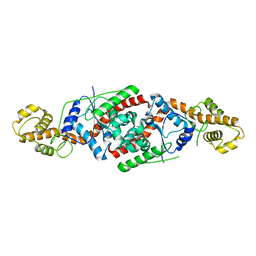 | | tryptophanyl-tRNA synthetase Val144Pro mutant from B. subtilis | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Antonczak, A.K, Simova, Z, Yonemoto, I, Bochtler, M, Piasecka, A, Czapinska, H, Brancale, A, Tippmann, E.M. | | Deposit date: | 2010-11-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of single molecular determinants in the fidelity of expanded genetic codes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FF1
 
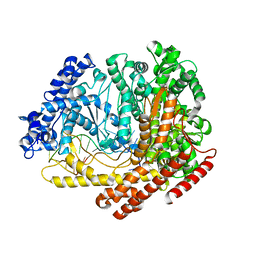 | |
4FF4
 
 | |
6OWL
 
 | | RNA oligonucleotides with 3'-arabino guanosine co-crystallized with GMP | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*R)-3'), RNA (5'-R(P*G)-3') | | Authors: | Szostak, J.W, Kim, S, Zhang, W. | | Deposit date: | 2019-05-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Model for the Emergence of RNA from a Prebiotically Plausible Mixture of Ribonucleotides, Arabinonucleotides, and 2'-Deoxynucleotides.
J.Am.Chem.Soc., 142, 2020
|
|
3L25
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4FF3
 
 | | N4 mini-vRNAP transcription initiation complex, 3 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
4H5O
 
 | |
1J7G
 
 | |
