5SCE
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 55 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCH
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 100 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCF
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 99 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)glycine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SC8
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 17 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-[(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)amino]pentanoic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
4KAO
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 1-(5-tert-Butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-(4-pyridin-3- yl-phenyl)-urea | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(pyridin-3-yl)phenyl]urea, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Graedler, U, Heinrich, T, Lehmann, M, Dresing, V. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of focal adhesion kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1URN
 
 | | U1A MUTANT/RNA COMPLEX + GLYCEROL | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTEIN (U1A), ... | | Authors: | Oubridge, C, Ito, N, Evans, P.R, Teo, C.-H, Nagai, K. | | Deposit date: | 1995-01-04 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin.
Nature, 372, 1994
|
|
1DIM
 
 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM COMPLEXED WITH EPANA INHIBITOR | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, POTASSIUM ION, SIALIDASE | | Authors: | Garman, E.F, Crennell, S.C, Vimr, E.R, Laver, W.G, Taylor, G.L. | | Deposit date: | 1996-04-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
1DJS
 
 | |
1V3X
 
 | | Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine | | Descriptor: | (2R)-4-[(6-CHLORO-2-NAPHTHYL)SULFONYL]-1-[(5-METHYL-4,5,6,7-TETRAHYDRO[1,3]THIAZOLO[5,4-C]PYRIDIN-2-YL)CARBONYL]PIPERAZ INE-2-CARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element
J.Med.Chem., 47, 2004
|
|
1R81
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate-N-acetyl-galactose | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1VAK
 
 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)65 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumer, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-18 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
4IE1
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 1h | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-8-hydroxyoctyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1VB4
 
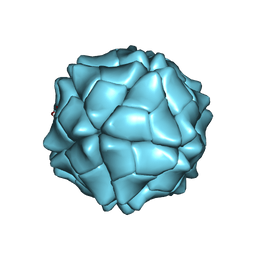 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)36 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1HPK
 
 | |
3QS8
 
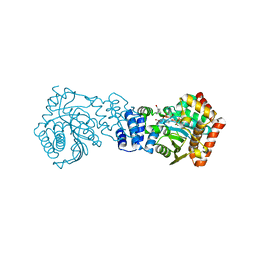 | | Anthranilate phosphoribosyltransferase (trpD) from Mycobacterium tuberculosis (complex with inhibitor ACS174) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-benzylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-02-20 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition.
Biochemistry, 52, 2013
|
|
3VII
 
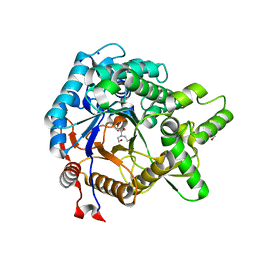 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
4FW4
 
 | | Crystal Structure of the LpxC in complex with N-[(1S,2R)-2-HYDROXY-1-(HYDROXYCARBAMOYL)PROPYL]-4-(4-PHENYLBUTA-1,3-DIYN-1-YL)BENZAMIDE inhibitor | | Descriptor: | ACETATE ION, GLYCEROL, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2012-06-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the LpxC
to be published
|
|
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
2E6O
 
 | |
4IME
 
 | |
2EHF
 
 | |
4G8H
 
 | | Crystal structure of C-lobe of bovine lactoferrin complexed with licofelone at 1.88 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of C-lobe of bovine lactoferrin complexed with licofelone at 1.88 A resolution
To be Published
|
|
4FW5
 
 | |
3R9P
 
 | |
