2C9G
 
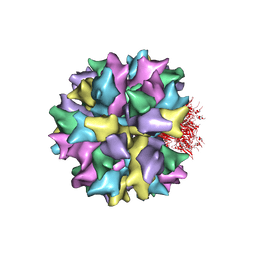 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON BASE DODECAHEDRON | | Descriptor: | PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
2C9H
 
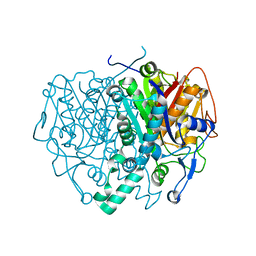 | | Structure of mitochondrial beta-ketoacyl synthase | | Descriptor: | MITOCHONDRIAL BETA-KETOACYL SYNTHASE, NICKEL (II) ION | | Authors: | Bunkoczi, G, Wu, X, Smee, C, Gileadi, O, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Oppermann, U. | | Deposit date: | 2005-12-12 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Mitochondrial Beta-Ketoacyl Synthase
To be Published
|
|
2C9I
 
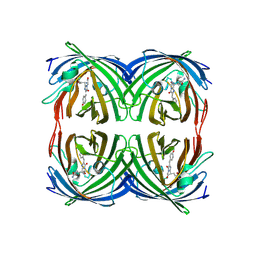 | | Structure of the fluorescent protein asFP499 from Anemonia sulcata | | Descriptor: | GREEN FLUORESCENT PROTEIN ASFP499 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chromophore-Protein Interactions in the Anthozoan Green Fluorescent Protein Asfp499
Biophys.J., 91, 2006
|
|
2C9J
 
 | | Structure of the fluorescent protein cmFP512 at 1.35A from Cerianthus membranaceus | | Descriptor: | GREEN FLUORESCENT PROTEIN FP512 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2006-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploring Chromophore-Protein Interactions in Fluorescent Protein Cmfp512 from Cerianthus Membranaceus: X-Ray Structure Analysis and Optical Spectroscopy.
Biochemistry, 45, 2006
|
|
2C9K
 
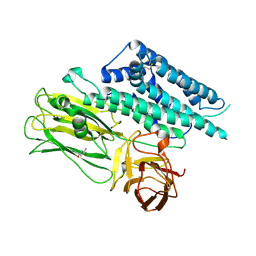 | | Structure of the functional form of the mosquito-larvicidal Cry4Aa toxin from Bacillus thuringiensis at 2.8 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PESTICIDAL CRYSTAL PROTEIN CRY4AA | | Authors: | Boonserm, P, Mo, M, Angsuthanasombat, C, Lescar, J. | | Deposit date: | 2005-12-13 | | Release date: | 2006-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Functional Form of the Mosquito Larvicidal Cry4Aa Toxin from Bacillus Thuringiensis at 2.8-Angstrom Resolution.
J.Bacteriol., 188, 2006
|
|
2C9L
 
 | | Structure of the Epstein-Barr virus ZEBRA protein | | Descriptor: | 5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*CP *TP*CP*AP*TP*GP*AP*AP*GP*T)-3', 5'-D(*AP*CP*TP*TP*CP*AP*CP*TP*GP*AP *GP*TP*CP*AP*GP*TP*GP*CP*T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
2C9M
 
 | | Structure of (SR) Calcium-ATPase in the Ca2E1 state solved in a P1 crystal form. | | Descriptor: | CALCIUM ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Lund Jensen, A.-M, Sorensen, T.L.-M, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Modulatory and Catalytic Modes of ATP Binding by the Calcium Pump
Embo J., 25, 2006
|
|
2C9N
 
 | | Structure of the Epstein-Barr virus ZEBRA protein at approximately 3. 5 Angstrom resolution | | Descriptor: | 5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP *T)-3', 5'-D(*CP*AP*TP*GP*AP*GP*TP*CP*AP*GP *T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
2C9O
 
 | | 3D Structure of the human RuvB-like helicase RuvBL1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RUVB-LIKE 1 | | Authors: | Matias, P.M, Gorynia, S, Donner, P, Carrondo, M.A. | | Deposit date: | 2005-12-14 | | Release date: | 2006-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human AAA+ protein RuvBL1.
J. Biol. Chem., 281, 2006
|
|
2C9P
 
 | | Cu(I)Cu(II)-CopC at pH 4.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C, NITRATE ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9Q
 
 | | Cu(I)Cu(II)-CopC at pH 7.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9R
 
 | | apo-H91F CopC | | Descriptor: | COPPER RESISTANCE PROTEIN C, SODIUM ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9S
 
 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9T
 
 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2C9U
 
 | | 1.24 Angstroms resolution structure of as-isolated Cu-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9V
 
 | | Atomic resolution structure of Cu-Zn Human Superoxide dismutase | | Descriptor: | COPPER (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9W
 
 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
2C9X
 
 | | Sulfite dehydrogenase from Starkeya Novella Y236F mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P, Tollin, G, Enemark, J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Evidence for the Importance of Tyr236 for the Integrity of the Mo Active Site in a Bacterial Sulfite Dehydrogenase.
Biochemistry, 45, 2006
|
|
2C9Y
 
 | | Structure of human adenylate kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE KINASE ISOENZYME 2, MITOCHONDRIAL, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Debreczeni, J.E, Turnbull, A, Papagrigoriou, E, Savitsky, P, Colebrook, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human Adenylate Kinase 2
To be Published
|
|
2C9Z
 
 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2CA0
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Yc-17-Bound Cytochrome P450 Pikc (Cyp107L1)
To be Published
|
|
2CA1
 
 | | Crystal structure of the IBV coronavirus nucleocapsid | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collison, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
2CA2
 
 | | CRYSTALLOGRAPHIC STUDIES OF INHIBITOR BINDING SITES IN HUMAN CARBONIC ANHYDRASE II. A PENTACOORDINATED BINDING OF THE SCN-ION TO THE ZINC AT HIGH P*H | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, THIOCYANATE ION, ... | | Authors: | Eriksson, A.E, Kylsten, P.M, Jones, T.A, Liljas, A. | | Deposit date: | 1989-02-06 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH.
Proteins, 4, 1988
|
|
2CA3
 
 | | Sulfite dehydrogenase from Starkeya Novella r55m mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P.V, Tollin, G, Enemark, J.H. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
J.Biol.Chem., 284, 2009
|
|
2CA4
 
 | | Sulfite dehydrogenase from Starkeya Novella mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFITE:CYTOCHROME C OXIDOREDUCTASE SUBUNIT A, ... | | Authors: | Bailey, S, Kappler, U. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis for Enzymatic Sulfite Oxidation: How Three Conserved Active Site Residues Shape Enzyme Activity.
J.Biol.Chem., 284, 2009
|
|
