1Z1C
 
 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute virus of Mice | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*AP*CP*AP*CP*CP*AP*AP*AP*A)-3', 5'-D(*AP*TP*CP*CP*TP*CP*TP*AP*TP*CP*AP*C)-3', ... | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
1U8S
 
 | |
5WSG
 
 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | Descriptor: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5WA1
 
 | | CHMP4C A232T in complex with ALIX BRO1 | | Descriptor: | Charged multivesicular body protein 4c, Programmed cell death 6-interacting protein | | Authors: | Wenzel, D.M, Alam, S.L, Sundquist, W.I. | | Deposit date: | 2017-06-24 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1TZT
 
 | | T. maritima NusB, P21 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
4MJO
 
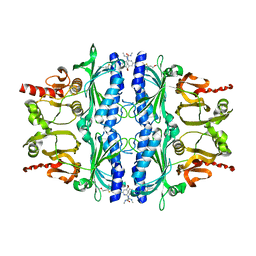 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
7SXI
 
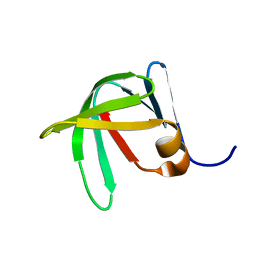 | | Solution Structure of Sds3 Capped Tudor Domain | | Descriptor: | Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Marcum, R.D, Radhakrishnan, I. | | Deposit date: | 2021-11-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A capped Tudor domain within a core subunit of the Sin3L/Rpd3L histone deacetylase complex binds to nucleic acid G-quadruplexes.
J.Biol.Chem., 298, 2022
|
|
1M7W
 
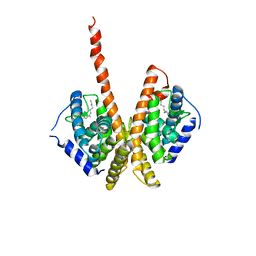 | | HNF4a ligand binding domain with bound fatty acid | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID | | Authors: | Dhe-Paganon, S, Duda, K, Iwamoto, M, Chi, Y.I, Shoelson, S.E. | | Deposit date: | 2002-07-22 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand
J.Biol.Chem., 277, 2002
|
|
2LD1
 
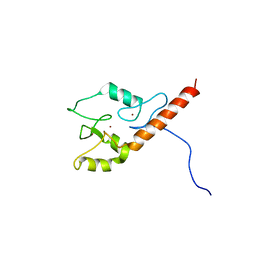 | |
6KFP
 
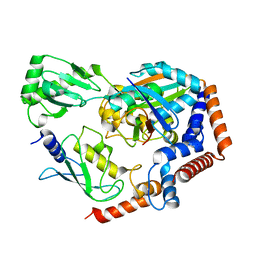 | | Crystal structure of MavC ternary complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
1TZX
 
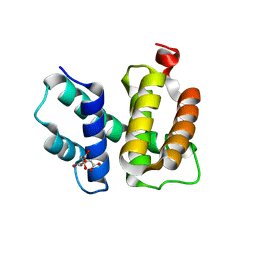 | | T. maritima NusB, P3221 | | Descriptor: | CITRIC ACID, N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
7TGG
 
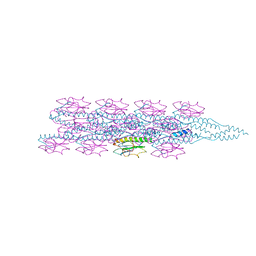 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
8FF8
 
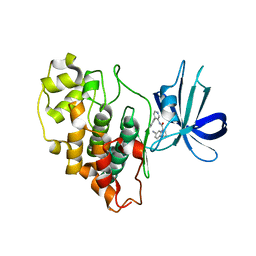 | |
1M1E
 
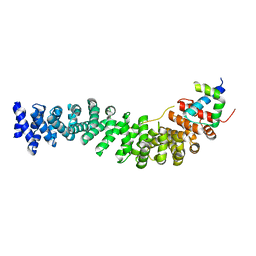 | | Beta-catenin armadillo repeat domain bound to ICAT | | Descriptor: | Beta-catenin, ICAT | | Authors: | Daniels, D.L, Weis, W.I. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ICAT inhibits Beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules.
Mol.Cell, 10, 2002
|
|
5X0Q
 
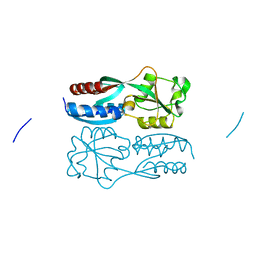 | | OxyR2 E204G variant (Cl-bound) from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, CITRIC ACID, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2017-01-23 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
5WHG
 
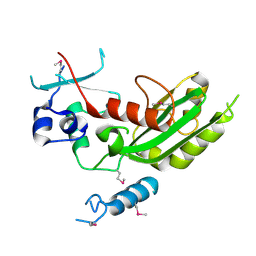 | | Vms1 mitochondrial localization core | | Descriptor: | Protein VMS1, ZINC ION | | Authors: | Fredrickson, E.K, Schubert, H.L, Rutter, J, Hill, C.P. | | Deposit date: | 2017-07-17 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sterol Oxidation Mediates Stress-Responsive Vms1 Translocation to Mitochondria.
Mol. Cell, 68, 2017
|
|
1ZZ6
 
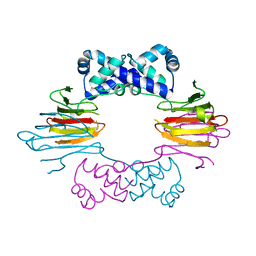 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
8CW4
 
 | | CryoEM structure of the N-pilus from Escherichia coli | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Conjugal transfer protein TraM | | Authors: | Bui, K.H, Black, C.S. | | Deposit date: | 2022-05-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T-pilus reveals the importance of positive charges in the lumen.
Structure, 31, 2023
|
|
8CUE
 
 | | CryoEM structure of the T-pilus from Agrobacterium tumefaciens | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Protein virB2 | | Authors: | Bui, K.H, Black, C.S. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T-pilus reveals the importance of positive charges in the lumen.
Structure, 31, 2023
|
|
8IYD
 
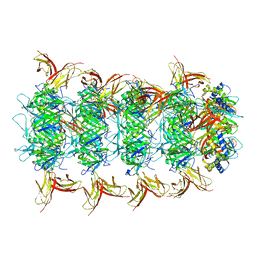 | | Tail cap of phage lambda tail | | Descriptor: | Tail tube protein, Tail tube terminator protein | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYK
 
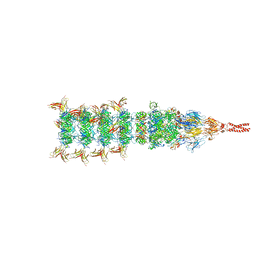 | | Tail tip conformation 1 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYL
 
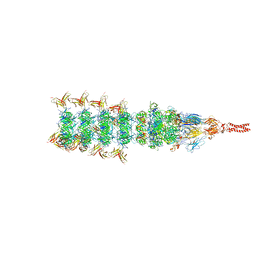 | | Tail tip conformation 2 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
1ZZ7
 
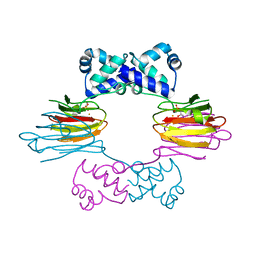 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
8FXP
 
 | | Structure of capsid of Agrobacterium phage Milano | | Descriptor: | Linking protein 1, gp16, Linking protein 2, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
1ZZ9
 
 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
