4V8E
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V97
 
 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2012-04-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7Y
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V9P
 
 | |
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
4V94
 
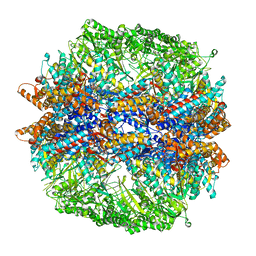 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
4V4F
 
 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a RNA molecule containing UAGAU repeats | | Descriptor: | 5'-R(*UP*AP*GP*AP*UP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
4V9F
 
 | |
4V5F
 
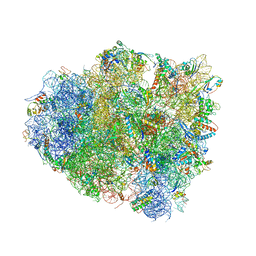 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V6M
 
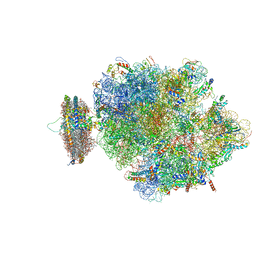 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5HQA
 
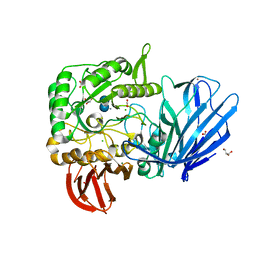 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
3BC9
 
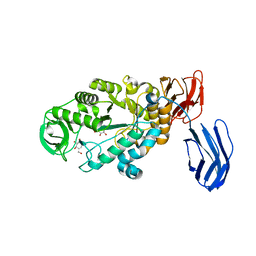 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
2ZQ0
 
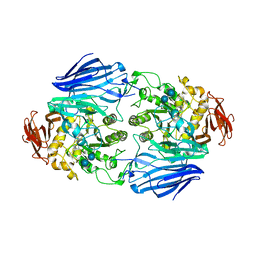 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
3AIC
 
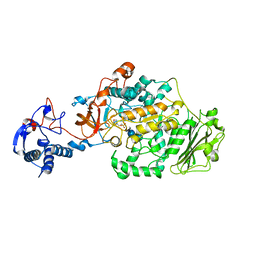 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
4N9O
 
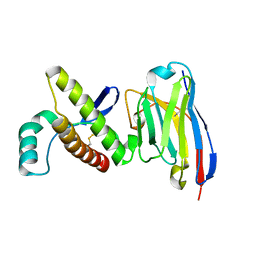 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4LIP
 
 | |
8C0O
 
 | |
4NX0
 
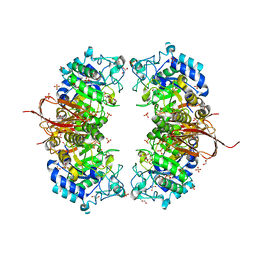 | | Crystal structure of Abp-WT, a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus | | Descriptor: | Abp, a GH27 beta-L-arabinopyranosidase, CITRIC ACID, ... | | Authors: | Lansky, S, Solomon, H.V, Salama, R, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-specificity relationships in Abp, a GH27 beta-L-arabinopyranosidase from Geobacillus stearothermophilus T6
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TTG
 
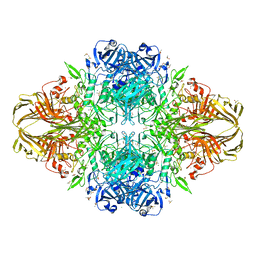 | | Beta-galactosidase (E. coli) in the presence of potassium chloride. | | Descriptor: | Beta-galactosidase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2014-06-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating factors important for monovalent cation selectivity in enzymes: E. coli beta-galactosidase as a model.
Phys Chem Chem Phys, 17, 2015
|
|
8HTU
 
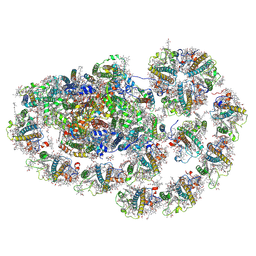 | | Cryo-EM structure of PpPSI-L | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Li, M, Pan, X.W, Sun, H.Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
8IMK
 
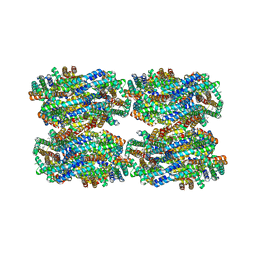 | | D3-D4, D1-D2, D'3-D'4, D'1-D'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster C) | | Descriptor: | ApcA1, ApcA2, ApcB1, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMI
 
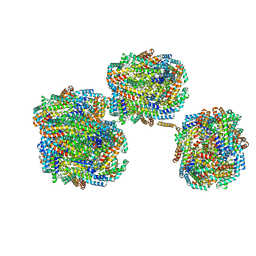 | | A1-A2, A3-A4, B'1-B'2, C'1-C'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster A) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMJ
 
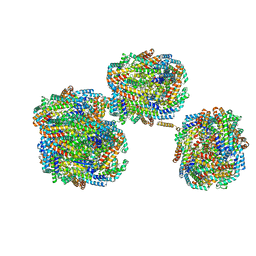 | | A'1-A'2, A'3-A'4, B1-B2, C1-C2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster B) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8I7O
 
 | | In situ structure of axonemal doublet microtubules in mouse sperm with 16-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 276, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
