2BDS
 
 | |
1RDG
 
 | |
4MDH
 
 | |
1HOE
 
 | |
5CYT
 
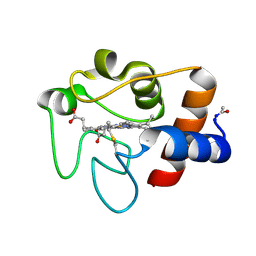 | | REFINEMENT OF MYOGLOBIN AND CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Takano, T. | | Deposit date: | 1988-01-14 | | Release date: | 1989-07-12 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refinement of Myoglobin and Cytochrome C
Methods and Applications in Crystallographic Computing, 1984
|
|
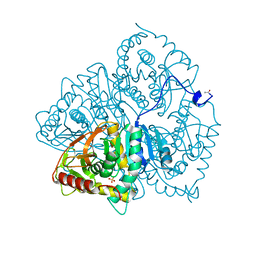
6LDH
 
 | |
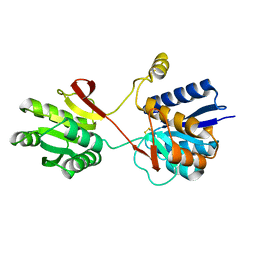
2LBP
 
 | |
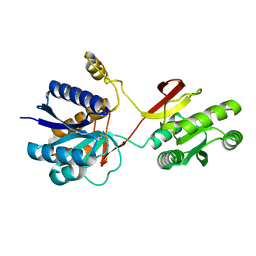
2LIV
 
 | |
3HFM
 
 | |
4GPD
 
 | |
2IG2
 
 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
1LDM
 
 | |
8LDH
 
 | |
1LLC
 
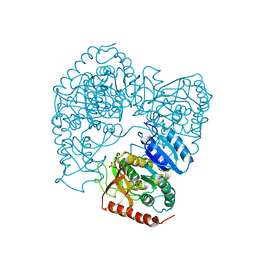 | | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS CASEI AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, L-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Buehner, M, Hecht, H.J, Hensel, R. | | Deposit date: | 1988-11-21 | | Release date: | 1989-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS-CASEI AT 3A RESOLUTION.
Acta Crystallogr.,Sect.A, 40, 1984
|
|
1LDB
 
 | |
2LDB
 
 | |
2FB4
 
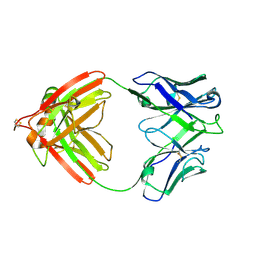 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
1BMV
 
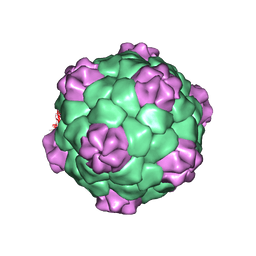 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
3TS1
 
 | |
1ALC
 
 | |
2RNT
 
 | | THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, GUANYLYL-2',5'-PHOSPHOGUANOSINE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Koepke, J, Maslowska, M, Heinemann, U. | | Deposit date: | 1988-07-06 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution.
J.Mol.Biol., 206, 1989
|
|
3HVP
 
 | |
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
2UTG
 
 | |
8DFR
 
 | | REFINED CRYSTAL STRUCTURES OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE. 3 ANGSTROMS APO-ENZYME AND 1.7 ANGSTROMS NADPH HOLO-ENZYME COMPLEX | | Descriptor: | CALCIUM ION, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Matthews, D.A, Oatley, S.J, Burridge, J, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined Crystal Structures of Chicken Liver Dihydrofolate Reductase. 3 Angstroms Apo-Enzyme and 1.7 Angstroms Nadph Holo-Enzyme Complex
To be Published
|
|
