4AQ0
 
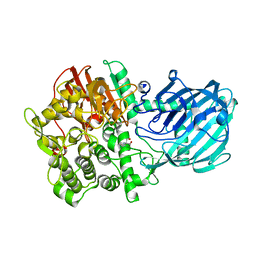 | | Structure of the Gh92 Family Glycosyl Hydrolase Ccman5 in complex with deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baranova, E, Tiels, P, Callewaert, N, Remaut, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Bacterial Glycosidase Enables Mannose-6-Phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes.
Nat.Biotechnol., 30, 2012
|
|
4GDZ
 
 | |
3CMZ
 
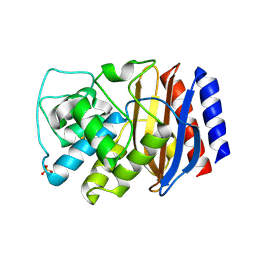 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
3RRQ
 
 | |
4ARK
 
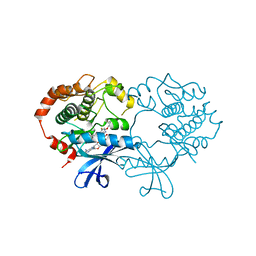 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN MAP KINASE KINASE 1 (MEK1) IN COMPLEX WITH A SMALL MOLECULE INHIBITOR AND ADP | | Descriptor: | 2-([3R-3,4-dihydroxybutyl]oxy)-4-fluoro-6-[(2-fluoro-4-iodophenyl)amino]benzamide, ADENOSINE-5'-DIPHOSPHATE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, ... | | Authors: | Hartung, I.V, Hitchcock, M, Puehler, F, Neuhaus, R, Scholz, A, Hammer, S, Petersen, K, Siemeister, G, Brittain, D, Hillig, R.C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Allosteric Mek Inhibitors - Part 1: Venturing Into Unexplored Sar Territories
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2H8A
 
 | |
3QK8
 
 | |
2WDB
 
 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
3CS7
 
 | | FACTOR XA IN COMPLEX WITH THE INHIBITOR 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one | | Descriptor: | 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one, CALCIUM ION, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Achieving structural diversity using the perpendicular conformation of alpha-substituted phenylcyclopropanes to mimic the bioactive conformation of ortho-substituted biphenyl P4 moieties: discovery of novel, highly potent inhibitors of Factor Xa.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1G4R
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN 1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
3D5P
 
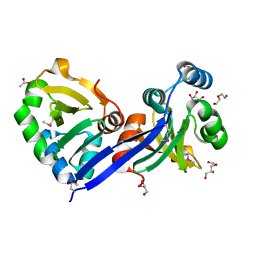 | |
3RL4
 
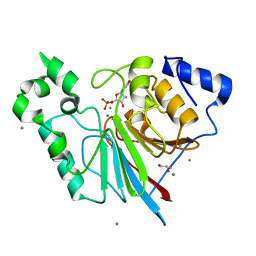 | | Rat metallophosphodiesterase MPPED2 G252H Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
1GIA
 
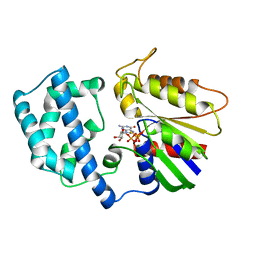 | |
4BRF
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with a distorted orthomolybdate ion and AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
3RAQ
 
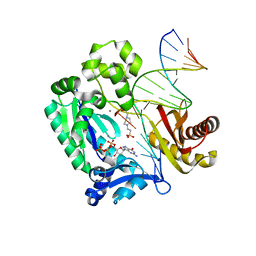 | |
2XA3
 
 | | crystal structure of the broadly neutralizing llama VHH D7 and its mode of HIV-1 gp120 interaction | | Descriptor: | LLAMA HEAVY CHAIN ANTIBODY D7, SULFATE ION | | Authors: | Hinz, A, Lutje Hulsik, D, Forsman, A, Koh, W, Belrhali, H, Gorlani, A, de Haard, H, Weiss, R.A, Verrips, T, Weissenhorn, W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the neutralizing Llama V(HH) D7 and its mode of HIV-1 gp120 interaction.
PLoS ONE, 5, 2010
|
|
4BTV
 
 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4CT3
 
 | | Methylmercury chloride derivative structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
1BAK
 
 | |
2UYU
 
 | |
3Q6Q
 
 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with disordered HBGA type Lea | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-01-03 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
4CPV
 
 | |
2H8S
 
 | | Solution structure of alpha-conotoxin Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J, Fischer, H, Nevin, S.T, Adams, D.J, Craik, D.J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The Synthesis, Structural Characterization, and Receptor Specificity of the {alpha}-Conotoxin Vc1.1.
J.Biol.Chem., 281, 2006
|
|
2WK7
 
 | | Structure of apo form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, SULFATE ION | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
1UNE
 
 | |
