5RGX
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1344037997 (Mpro-x2572) | | Descriptor: | 2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RJS
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z285642082 | | Descriptor: | N-cyclopropylpyrazolo[1,5-a]pyrimidine-3-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK9
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z461898648 | | Descriptor: | N,N-dimethyl-1H-pyrazole-4-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1545196101 | | Descriptor: | 2-fluoro-N-[(3-methyl-1H-pyrazol-4-yl)methyl]aniline, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJV
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z57190020 | | Descriptor: | PH-interacting protein, methyl {4-[(pyridin-4-yl)methyl]phenyl}carbamate | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKC
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z234898257 | | Descriptor: | N-methyl-1-([1,2,4]triazolo[4,3-a]pyridin-3-yl)methanamine, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKT
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1266933824 | | Descriptor: | (1H-pyrazol-4-yl)(pyrrolidin-1-yl)methanone, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RW0
 
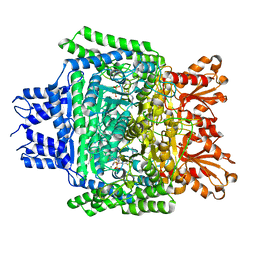 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z2444997446 | | Descriptor: | 1-{(2S,4S)-4-fluoro-1-[(2-methyl-1,3-thiazol-4-yl)methyl]pyrrolidin-2-yl}methanamine, MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RGW
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4444621910 (Mpro-x2569) | | Descriptor: | 2-(5-cyanopyridin-3-yl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RJO
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00023827 | | Descriptor: | 6-bromo-1,3-dihydro-2H-indol-2-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK1
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1507502062 | | Descriptor: | 6-methoxy-1,3,4,5-tetrahydro-2H-1-benzazepin-2-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKI
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 | | Descriptor: | 4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.268 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RGU
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4444622180 (Mpro-x2562) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RHA
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z147647874 (Mpro-x2779) | | Descriptor: | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RJQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024665 | | Descriptor: | 2-(4-bromanyl-2-methoxy-phenyl)ethanoic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK5
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z57478994 | | Descriptor: | 5-(methoxymethyl)-1,3,4-thiadiazol-2-amine, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
4MB0
 
 | | Crystal structure of TON1374 | | Descriptor: | ACETATE ION, phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
5SDE
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1619978933 | | Descriptor: | 5-fluoro-1-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]-1,2,3,6-tetrahydropyridine, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434854 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]piperidine-4-carboxamide, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDG
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z136583524 | | Descriptor: | 2-methyl-N-(pyridin-4-yl)furan-3-carboxamide, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDO
 
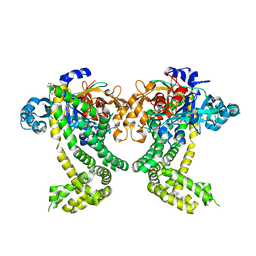 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z19735067 | | Descriptor: | 2-(4-fluorophenoxy)-1-(pyrrolidin-1-yl)ethan-1-one, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDQ
 
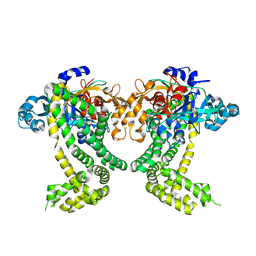 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434887 | | Descriptor: | 2-[(morpholin-4-yl)methyl]phenol, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDC
 
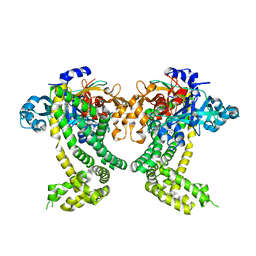 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434912 | | Descriptor: | 3-[(4-methylpiperidin-1-yl)methyl]-1H-indole, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDI
 
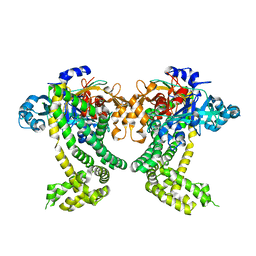 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z44592329 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-phenyl-N'-pyridin-3-ylurea | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDL
 
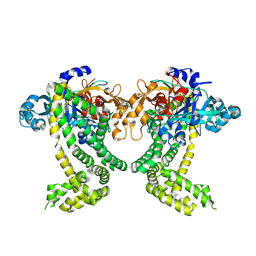 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z321318226 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
