3PB2
 
 | |
4X1T
 
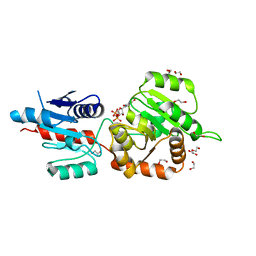 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
7FSE
 
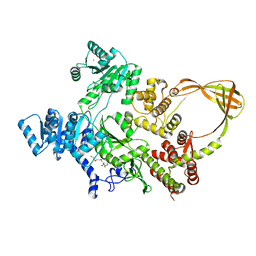 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch | | Descriptor: | CHLORIDE ION, DODECAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7FSF
 
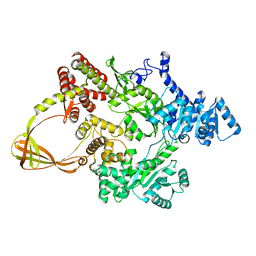 | | CRYSTAL STRUCTURE OF T. MARITIMA REVERSE GYRASE ACTIVE SITE VARIANT Y851F | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5IPW
 
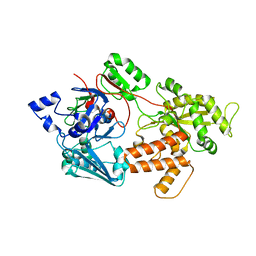 | | oligopeptide-binding protein OppA | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, putative | | Authors: | Lee, H.H, Kim, H.J, Yoon, H.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a putative oligopeptide-binding periplasmic protein from a hyperthermophile
Extremophiles, 20, 2016
|
|
5H72
 
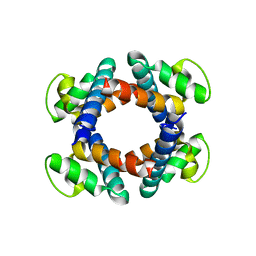 | | Structure of the periplasmic domain of FliP | | Descriptor: | Flagellar biosynthetic protein FliP | | Authors: | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-11-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
5HM4
 
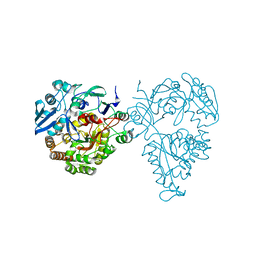 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
1Q7Q
 
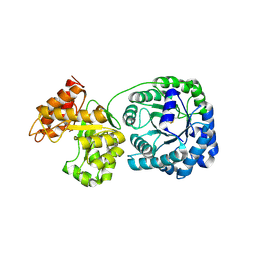 | | Cobalamin-dependent methionine synthase (1-566) from T. maritima (Oxidized, Orthorhombic) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-19 | | Release date: | 2004-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3QF4
 
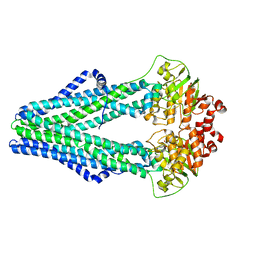 | | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hohl, M, Briand, C, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QF7
 
 | |
3QG5
 
 | |
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S, Myles, D.A. | | Deposit date: | 2022-06-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
4EED
 
 | | CorA coiled-coil mutant under Mg2+ presence | | Descriptor: | MAGNESIUM ION, Magnesium transport protein CorA | | Authors: | Pfoh, R, Pai, E.F. | | Deposit date: | 2012-03-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structural asymmetry in the magnesium channel CorA points to sequential allosteric regulation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EEB
 
 | | CorA coiled-coil mutant under Mg2+ absence | | Descriptor: | CESIUM ION, Magnesium transport protein CorA, SODIUM ION | | Authors: | Pfoh, R, Pai, E.F. | | Deposit date: | 2012-03-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural asymmetry in the magnesium channel CorA points to sequential allosteric regulation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6F4L
 
 | |
1X9J
 
 | |
6G74
 
 | |
4B20
 
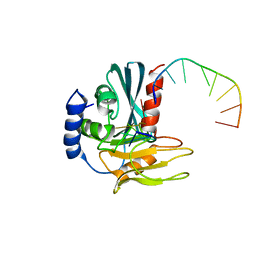 | | Structural basis of DNA loop recognition by Endonuclease V | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*AP*CP*AP*GP)-3', ENDONUCLEASE V, ... | | Authors: | Rosnes, I, Rowe, A.D, Forstrom, R.J, Alseth, I, Bjoras, M, Dalhus, B. | | Deposit date: | 2012-07-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of DNA Loop Recognition by Endonuclease V.
Structure, 21, 2013
|
|
3IH2
 
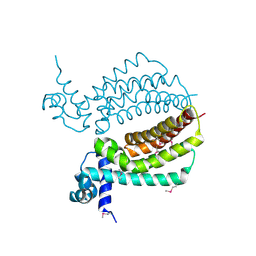 | | TM1030 crystallized at 323K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
3IH4
 
 | | TM1030 crystallized at 277K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
3IH3
 
 | | TM1030 crystallized at 310K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
7WYT
 
 | | Crystal structures of Na+,K+-ATPase in complex with ouabain | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WYS
 
 | | Crystal structures of Na+,K+-ATPase in complex with istaroxime | | Descriptor: | (3E,5S,8R,9S,10R,13S,14S)-3-(2-azanylethoxyimino)-10,13-dimethyl-1,2,4,5,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthrene-6,17-dione, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Cryoelectron microscopy of Na + ,K + -ATPase in the two E2P states with and without cardiotonic steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2CH4
 
 | |
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
