1NVF
 
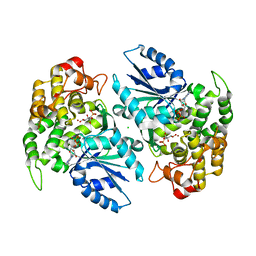 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, ADP and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
4R1M
 
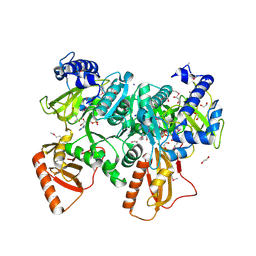 | |
1NVE
 
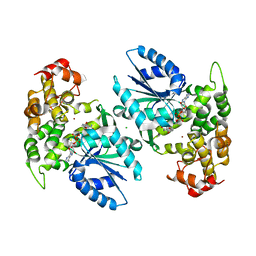 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1VWP
 
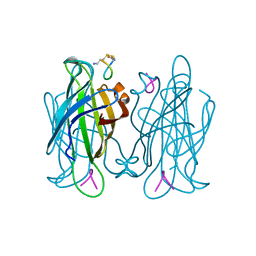 | |
1VWJ
 
 | |
2YZN
 
 | |
3NIL
 
 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
2QIZ
 
 | |
3NIK
 
 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
7BEU
 
 | | Human glutathione transferase M1-1 | | Descriptor: | Glutathione S-transferase Mu 1, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2020-12-28 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Ligandability Assessment of Human Glutathione Transferase M1-1 Using Pesticides as Chemical Probes.
Int J Mol Sci, 23, 2022
|
|
2YZM
 
 | |
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1SWV
 
 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
3NIH
 
 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
2YZG
 
 | |
1VWH
 
 | |
1VWE
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 3.6 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWL
 
 | |
1VWM
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.2 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1VWQ
 
 | |
