1ULP
 
 | |
1RDH
 
 | |
1RN1
 
 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
1TK9
 
 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
7OZJ
 
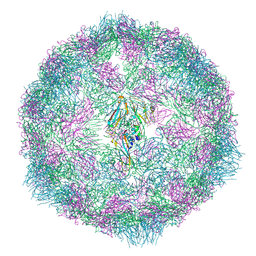 | |
7OZI
 
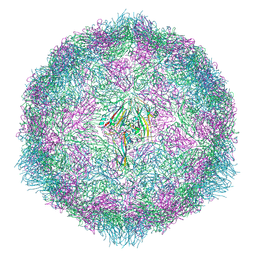 | |
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VAR
 
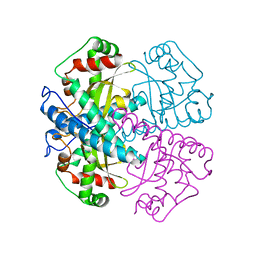 | |
1NTE
 
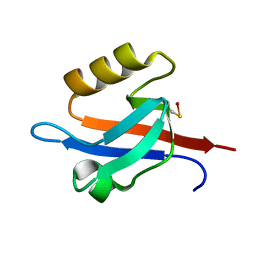 | | CRYSTAL STRUCTURE ANALYSIS OF THE SECOND PDZ DOMAIN OF SYNTENIN | | Descriptor: | OXYGEN ATOM, Syntenin 1 | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-29 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular roots of degenerate specificity in syntenin's PDZ2 domain: reassessment of the PDZ recognition paradigm.
Structure, 11, 2003
|
|
1NXB
 
 | |
7OZK
 
 | | CryoEM structure of human enterovirus 70 in complex with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7OZL
 
 | | CryoEM structure of human enterovirus 70 in complex with WIN51711 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
1O62
 
 | |
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1USN
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-142372 | | Descriptor: | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2YL)-UREIDO]-N-METHYL-3-PENTAFLUOROPHENYL-PROPIONAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Finzel, B.C, Bryant Junior, G.L, Baldwin, E.T. | | Deposit date: | 1998-06-09 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity.
Protein Sci., 7, 1998
|
|
1MVL
 
 | | PPC decarboxylase mutant C175S | | Descriptor: | FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
5CRO
 
 | |
5T5A
 
 | |
1TDE
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
1UXB
 
 | | ADENOVIRUS AD19p FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
1NPO
 
 | |
4OQF
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIB-SR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
1UXA
 
 | | ADENOVIRUS AD37 FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
5AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
