1D3R
 
 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
7AQS
 
 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
1CG6
 
 | |
1CGQ
 
 | |
1D5Z
 
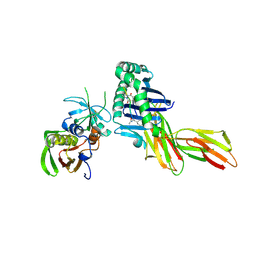 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | PROTEIN (ENTEROTOXIN TYPE B), PROTEIN (HLA CLASS II HISTOCOMPATIBILITY ANTIGEN), PROTEIN (PEPTIDOMIMETIC INHIBITOR) | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
5MZ8
 
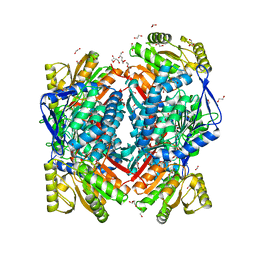 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5GMK
 
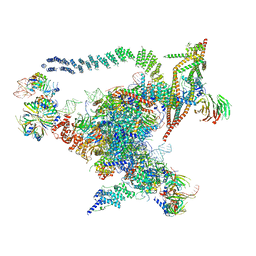 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
1D6P
 
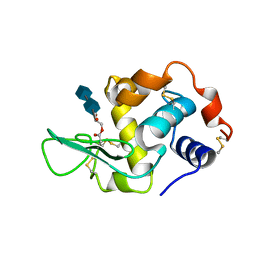 | | HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LYSOZYME | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
1CK4
 
 | | CRYSTAL STRUCTURE OF RAT A1B1 INTEGRIN I-DOMAIN. | | Descriptor: | INTEGRIN ALPHA-1 | | Authors: | Nolte, M, Pepinsky, R.B, Venyaminov, S.Y, Koteliansky, V, Gotwals, P.J, Karpusas, M. | | Deposit date: | 1999-04-27 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the alpha1beta1 integrin I-domain: insights into integrin I-domain function.
FEBS Lett., 452, 1999
|
|
1CKS
 
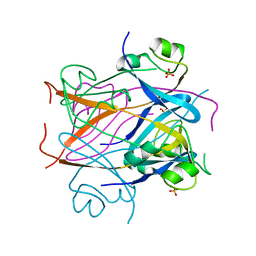 | |
1CMV
 
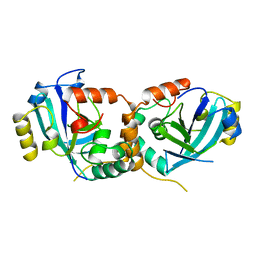 | | HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE | | Authors: | Shieh, H.-S, Kurumbail, R.G, Stevens, A.M, Stegeman, R.A, Sturman, E.J, Pak, J.Y, Wittwer, A.J, Palmier, M.O, Wiegand, R.C, Holwerda, B.C, Stallings, W.C. | | Deposit date: | 1996-08-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three-dimensional structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
1D8U
 
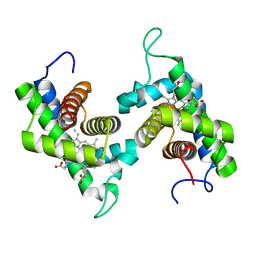 | | CRYSTAL STRUCTURE OF NON-SYMBIOTIC PLANT HEMOGLOBIN FROM RICE | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, M, Brucker, E.A, Stec, B, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1999-10-26 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a nonsymbiotic plant hemoglobin.
Structure Fold.Des., 8, 2000
|
|
5GKD
 
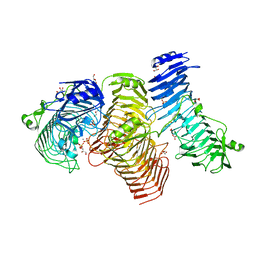 | | Structure of PL6 family alginate lyase AlyGC | | Descriptor: | AlyGC, CALCIUM ION, CARBONATE ION, ... | | Authors: | Zhang, Y.Z, Wang, P, Xu, F. | | Deposit date: | 2016-07-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6
J. Biol. Chem., 292, 2017
|
|
1CQY
 
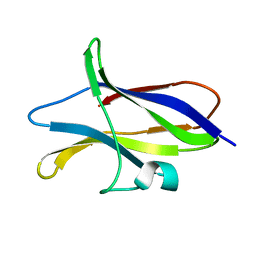 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
1CWN
 
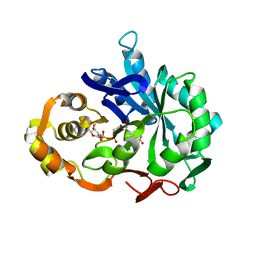 | | CRYSTAL STRUCTURE OF PORCINE ALDEHYDE REDUCTASE HOLOENZYME | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O. | | Deposit date: | 1996-07-30 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of porcine aldehyde reductase at 2.0 angstrom resolution: Modeling an inhibitor in the active site of the enzyme.
Protein Pept.Lett., 3, 1996
|
|
1CNE
 
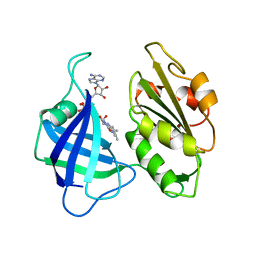 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
5GM1
 
 | | Crystal structure of methyltransferase TleD complexed with SAH | | Descriptor: | O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
5MZN
 
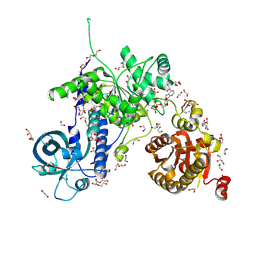 | | Helicase Sen1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leonaite, B, Basquin, J, Conti, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family.
EMBO J., 36, 2017
|
|
1CER
 
 | |
1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
1C50
 
 | | IDENTIFICATION AND STRUCTURAL CHARACTERIZATION OF A NOVEL ALLOSTERIC BINDING SITE OF GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Skamnaki, V.T, Tsitsanou, K.E, Gavalas, N.G, Johnson, L.N. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new allosteric site in glycogen phosphorylase b as a target for drug interactions.
Structure Fold.Des., 8, 2000
|
|
1BYD
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1C52
 
 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1C57
 
 | | DIRECT DETERMINATION OF THE POSITIONS OF DEUTERIUM ATOMS OF BOUND WATER IN CONCANAVALIN A BY NEUTRON LAUE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, Concanavalin-Br, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb, A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C4X
 
 | | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE (BPHD) FROM RHODOCOCCUS SP. STRAIN RHA1 | | Descriptor: | PROTEIN (2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE) | | Authors: | Nandhagopal, N, Senda, T, Mitsui, Y. | | Deposit date: | 1999-09-30 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway
Proc.Jpn.Acad.,Ser.B, 73, 1997
|
|
