7P9N
 
 | | Crystal structure of CD73 in complex with AMP in the open form | | Descriptor: | 5'-nucleotidase, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7P9R
 
 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PCP
 
 | |
7PBY
 
 | |
2YGX
 
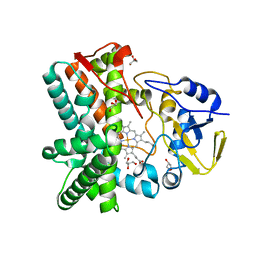 | | Structure of the mixed-function P450 MycG in P21 space group | | Descriptor: | GLYCEROL, P-450-LIKE PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-04-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
6TWA
 
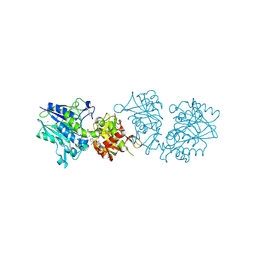 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12646 (an AOPCP derivative, compound 20 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-19 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
6A7J
 
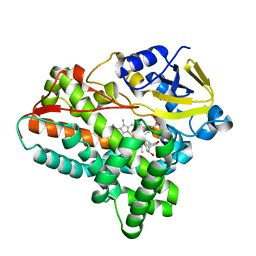 | | Testerone bound CYP154C4 from Streptomyces sp. ATCC 11861 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6WZP
 
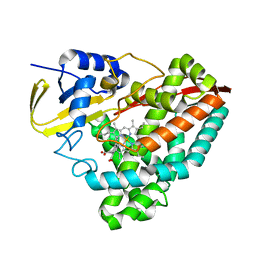 | | The crystal structure of 4-vinylbenzoate-bound T252A mutant CYP199A4 | | Descriptor: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
2YJN
 
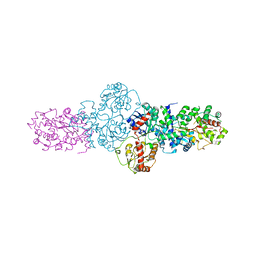 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | Descriptor: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | Authors: | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
6UNN
 
 | |
3L62
 
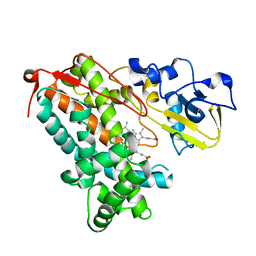 | | Crystal structure of substrate-free P450cam at low [K+] | | Descriptor: | Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
2Z3U
 
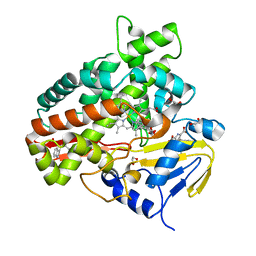 | | Crystal Structure of Chromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DI-1H-INDOL-3-YL-1H-PYRROLE-2,5-DICARBOXYLIC ACID, Cytochrome P450, ... | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Z3T
 
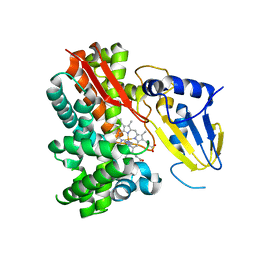 | | Crystal Structure of Substrate Free Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, IMIDAZOLE, ... | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6A7A
 
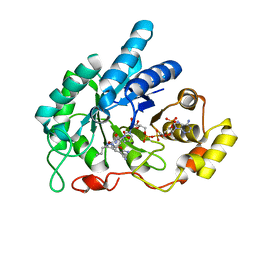 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6TW0
 
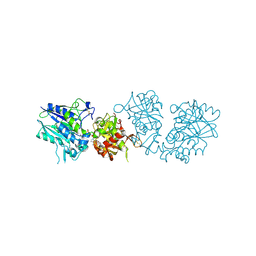 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
3VOO
 
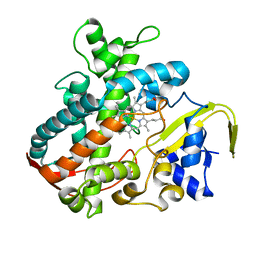 | | Cytochrome P450SP alpha (CYP152B1) mutant A245E | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
6TWF
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6XYT
 
 | | Crystal structure of the O-state of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
2Z36
 
 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
2ZAX
 
 | | Crystal Structure of Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Sakurai, K, Shimada, H, Harada, K, Hayashi, T, Tsukihara, T. | | Deposit date: | 2007-10-11 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evaluation of the functional role of the heme-6-propionate side chain in cytochrome P450cam
J.Am.Chem.Soc., 130, 2008
|
|
6TVX
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12676 (an AOPCP derivative, compound 9 in paper) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
5AZD
 
 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
6U30
 
 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
