1DTS
 
 | | CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE, DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-03-28 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 A resolution.
Structure, 2, 1994
|
|
1TPU
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
1FNB
 
 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FNC
 
 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FERREDOXIN-NADP+ REDUCTASE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FND
 
 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
1FCC
 
 | | CRYSTAL STRUCTURE OF THE C2 FRAGMENT OF STREPTOCOCCAL PROTEIN G IN COMPLEX WITH THE FC DOMAIN OF HUMAN IGG | | Descriptor: | IGG1 MO61 FC, STREPTOCOCCAL PROTEIN G (C2 FRAGMENT) | | Authors: | Sauer-Eriksson, A.E, Kleywegt, G.J, Uhlen, M, Jones, T.A. | | Deposit date: | 1995-01-17 | | Release date: | 1995-04-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG.
Structure, 3, 1995
|
|
1HTL
 
 | |
1BUC
 
 | | THREE-DIMENSIONAL STRUCTURE OF BUTYRYL-COA DEHYDROGENASE FROM MEGASPHAERA ELSDENII | | Descriptor: | ACETOACETYL-COENZYME A, BUTYRYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Djordjevic, S, Pace, C.P, Stankovich, M.T, Kim, J.J.P. | | Deposit date: | 1994-09-06 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii.
Biochemistry, 34, 1995
|
|
1TNM
 
 | |
1LPM
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPN
 
 | |
1PSF
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF PSAE FROM THE CYANOBACTERIUM SYNECHOCOCCUS SP. STRAIN PCC 7002: A PHOTOSYSTEM I PROTEIN THAT SHOWS STRUCTURAL HOMOLOGY WITH SH3 DOMAINS | | Descriptor: | PHOTOSYSTEM I ACCESSORY PROTEIN E | | Authors: | Falzone, C.J, Kao, Y.-H, Zhao, J, Bryant, D.A, Lecomte, J.T.J. | | Deposit date: | 1994-04-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of PsaE from the cyanobacterium Synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains.
Biochemistry, 33, 1994
|
|
1PSE
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF PSAE FROM THE CYANOBACTERIUM SYNECHOCOCCUS SP. STRAIN PCC 7002: A PHOTOSYSTEM I PROTEIN THAT SHOWS STRUCTURAL HOMOLOGY WITH SH3 DOMAINS | | Descriptor: | PHOTOSYSTEM I ACCESSORY PROTEIN E | | Authors: | Falzone, C.J, Kao, Y.-H, Zhao, J, Bryant, D.A, Lecomte, J.T.J. | | Deposit date: | 1994-04-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of PsaE from the cyanobacterium Synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains.
Biochemistry, 33, 1994
|
|
1EBG
 
 | |
107D
 
 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | Descriptor: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | Authors: | Lin, C.H, Patel, D.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
1TAD
 
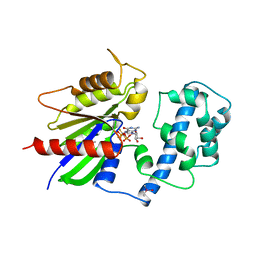 | | GTPASE MECHANISM OF GPROTEINS FROM THE 1.7-ANGSTROM CRYSTAL STRUCTURE OF TRANSDUCIN ALPHA-GDP-ALF4- | | Descriptor: | CACODYLATE ION, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sondek, J, Lambright, D.G, Noel, J.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1995-01-05 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4.
Nature, 372, 1994
|
|
1ASY
 
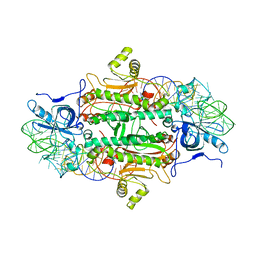 | | CLASS II AMINOACYL TRANSFER RNA SYNTHETASES: CRYSTAL STRUCTURE OF YEAST ASPARTYL-TRNA SYNTHETASE COMPLEXED WITH TRNA ASP | | Descriptor: | ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Ruff, M, Cavarelli, J, Rees, B, Krishnaswamy, S, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp).
Science, 252, 1991
|
|
1ITG
 
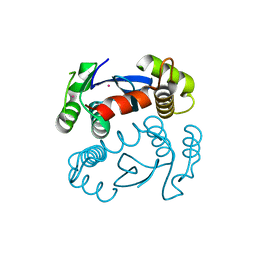 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
1LNF
 
 | |
1LND
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNB
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1HCG
 
 | |
1RSC
 
 | | STRUCTURE OF AN EFFECTOR INDUCED INACTIVATED STATE OF RIBULOSE BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE: THE BINARY COMPLEX BETWEEN ENZYME AND XYLULOSE BISPHOSPHATE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN), XYLULOSE-1,5-BISPHOSPHATE | | Authors: | Newman, J, Gutteridge, S. | | Deposit date: | 1994-03-29 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an effector-induced inactivated state of ribulose 1,5-bisphosphate carboxylase/oxygenase: the binary complex between enzyme and xylulose 1,5-bisphosphate.
Structure, 2, 1994
|
|
