7PB5
 
 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBA
 
 | | Crystal structure of CD73 in complex with IMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-08-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PA4
 
 | | Crystal structure of CD73 in complex with CMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBB
 
 | |
7P9N
 
 | | Crystal structure of CD73 in complex with AMP in the open form | | Descriptor: | 5'-nucleotidase, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7P9R
 
 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBY
 
 | |
8QZR
 
 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8QT5
 
 | |
8QTC
 
 | |
8QTT
 
 | |
8QTF
 
 | |
5L88
 
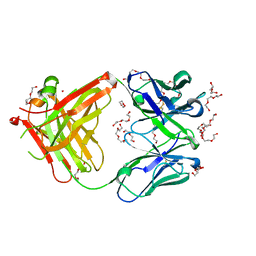 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L9D
 
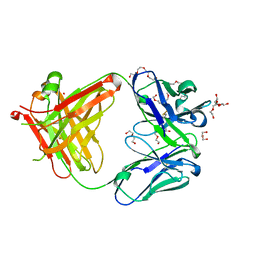 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSYLATED, CRYSTAL FORM I, parsimonious model | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-10 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4P4K
 
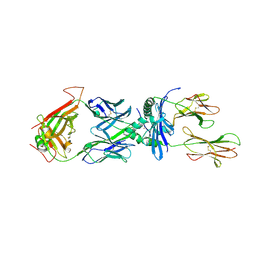 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between allergic hypersensitivity and auto immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM, HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
5L7X
 
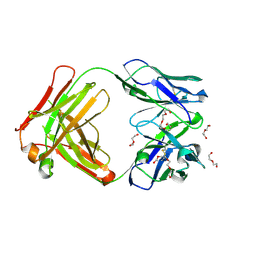 | | Afamin antibody fragment, N14 Fab, L1- glycosylated, crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Mouse Antibody Fab Fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
7YKJ
 
 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | Descriptor: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | Authors: | Tang, B, Dang, S. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
2A0A
 
 | |
119L
 
 | |
118L
 
 | |
120L
 
 | |
122L
 
 | |
123L
 
 | |
125L
 
 | |
