9MSZ
 
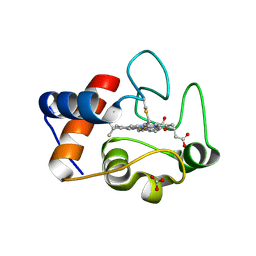 | |
3SJL
 
 | |
4W1V
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a thiazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
5CUL
 
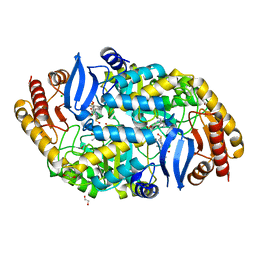
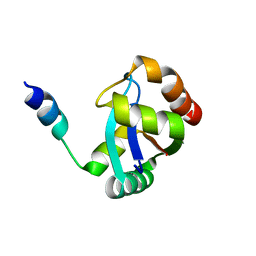 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
1LL9
 
 | | Crystal Structure Of AmpC beta-Lactamase From E. Coli In Complex With Amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, beta-lactamase | | Authors: | Trehan, I, Morandi, F, Blaszczak, L.C, Shoichet, B.K. | | Deposit date: | 2002-04-26 | | Release date: | 2002-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Using steric hindrance to design new inhibitors of class C beta-lactamases.
Chem.Biol., 9, 2002
|
|
9L48
 
 | | Crystal structure of HLA-C*12:02-RV9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Toll-like receptor 9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3SEO
 
 | |
8QS4
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 22 (1083853) | | Descriptor: | 14-3-3 protein sigma, C-RAF peptide pS259, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS7
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 70 (1084352) | | Descriptor: | 1-[8-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS6
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 21 (1075354) | | Descriptor: | 14-3-3 protein sigma, C-RAF peptide pS259, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS5
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 21 (1075354) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[3-fluoranyl-4-(trifluoromethyl)phenyl]sulfonylpiperidin-4-yl]methyl]ethanamide, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QSA
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 86 (1084384) | | Descriptor: | 1-[(5~{R})-2-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,7-diazaspiro[4.4]nonan-7-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS8
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 78 (1084378) | | Descriptor: | 1-[8-(4-bromophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS9
 
 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 83 (1084383) | | Descriptor: | 1-[8-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
9J0J
 
 | | The crystal structure of Fe/2OG-dependent oxygenase DfmD | | Descriptor: | FE (III) ION, Fe/2OG-dependent oxygenase DfmD, GLYCEROL, ... | | Authors: | Wang, L, Chen, J, Zhou, J. | | Deposit date: | 2024-08-02 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | An Alternative Mechanism for C-C Desaturation Underscores a Dual-Controlled Mechanism for the Fate of Radical Intermediate in Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenase DfmD.
J.Am.Chem.Soc., 147, 2025
|
|
5XGE
 
 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with cyclic di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein PA0861 | | Authors: | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
3SGD
 
 | | Crystal structure of the mouse mAb 17.2 | | Descriptor: | CALCIUM ION, Heavy Chain, Light Chain | | Authors: | Pizarro, J.C, Boulot, G, Hontebeyrie, M, Bentley, G.A. | | Deposit date: | 2011-06-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the complex mAb 17.2 and the C-terminal region of Trypanosoma cruzi P2 Beta protein: implications in cross-reactivity
Plos Negl Trop Dis, 5, 2011
|
|
9L4A
 
 | | Crystal structure of HLA-C*12:02-MY9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, MY9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
9L49
 
 | | Crystal structure of HLA-C*12:03-RV9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Toll-like receptor 9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
6EZI
 
 | |
9L47
 
 | | Crystal structure of HLA-C*12:03-MY9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, MY9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
3SGE
 
 | | Crystal structure of mAb 17.2 in complex with R13 peptide | | Descriptor: | CALCIUM ION, Heavy Chain, Light Chain, ... | | Authors: | Pizarro, J.C, Boulot, G, Hontebeyrie, M, Bentley, G.A. | | Deposit date: | 2011-06-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex mAb 17.2 and the C-terminal region of Trypanosoma cruzi P2 Beta protein: implications in cross-reactivity
Plos Negl Trop Dis, 5, 2011
|
|
4CWP
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
5DDT
 
 | |
