7NOD
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'4 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO3
 
 | | Structure of the mature RSV CA lattice: pentamer derived from polyhedral VLPs | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOH
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 5'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOF
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'4 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO9
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-pentamer interface, class 1'1 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOO
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Alpha | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO6
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NON
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO5
 
 | | Structure of the mature RSV CA lattice: hexamer with 2 adjacent pentamers (C2 symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOK
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO7
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"2 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOL
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Alpha | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO4
 
 | | Structure of the mature RSV CA lattice: hexamer with 3 adjacent pentamers (C3 symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOJ
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOC
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'3 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO1
 
 | | Structure of the mature RSV CA lattice: T=3 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOE
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NRI
 
 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NOP
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOA
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOQ
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NGQ
 
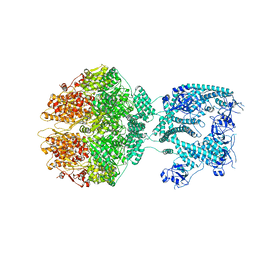 | | Human mitochondrial Lon protease homolog, D2-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Abrahams, J.P, Schmitz, K.A, Maier, T, Schenck, N. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGP
 
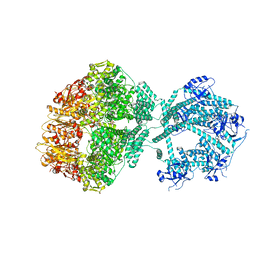 | | D1-state of wild type human mitochondrial LONP1 protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGF
 
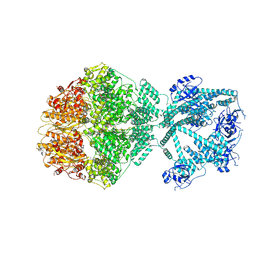 | | P2c-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGL
 
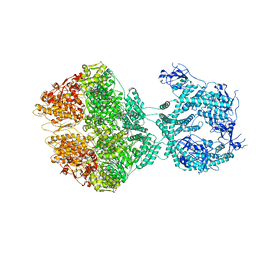 | | R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
