8SQX
 
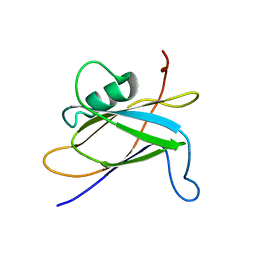 | |
4AV2
 
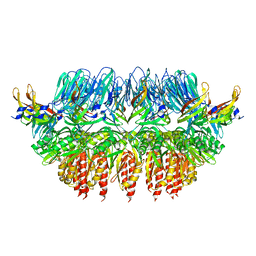 | | Single particle electron microscopy of PilQ dodecameric complexes from Neisseria meningitidis. | | Descriptor: | PILP PROTEIN, TYPE IV PILUS BIOGENESIS AND COMPETENCE PROTEIN PILQ | | Authors: | Berry, J.L, Phelan, M.M, Collins, R.F, Adomavicius, T, Tonjum, T, Frye, S.A, Bird, L, Owens, R, Ford, R.C, Lian, L.Y, Derrick, J.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Structure and Assembly of a Trans-Periplasmic Channel for Type Iv Pili in Neisseria Meningitidis.
Plos Pathog., 8, 2012
|
|
2N7H
 
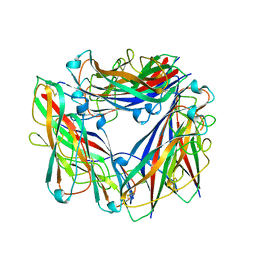 | | Hybrid structure of the Type 1 Pilus of Uropathogenic E.coli | | Descriptor: | FimA | | Authors: | Habenstein, B, Loquet, A, Giller, K, Vasa, S, Becker, S, Habeck, M, Lange, A. | | Deposit date: | 2015-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-14 | | Method: | SOLID-STATE NMR | | Cite: | Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
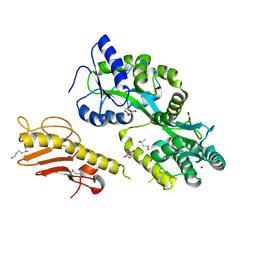
5VAW
 
 | |
3VOR
 
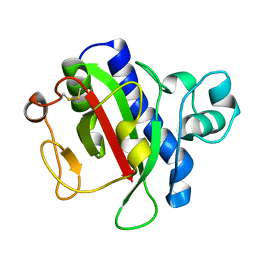 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5FGS
 
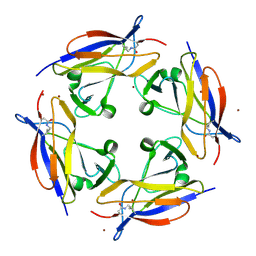 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group | | Descriptor: | Cell surface protein SpaA, ZINC ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
4DWH
 
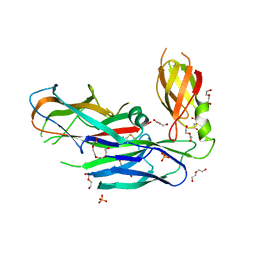 | | Structure of the major type 1 pilus subunit FIMA bound to the FIMC (2.5 A resolution) | | Descriptor: | Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Scharer, M.A, Puorger, C, Crespo, M, Glockshuber, R, Capitani, G. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
2X9W
 
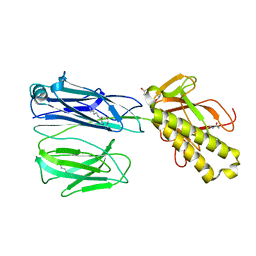 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
8DZF
 
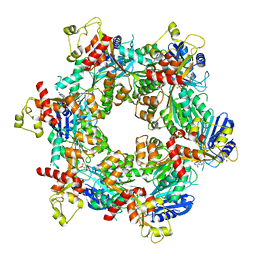 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of AMP-PNP (class-2) | | Descriptor: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZE
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E. coli in the presence of AMP-PNP (class-1) | | Descriptor: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZG
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BfpD, MAGNESIUM ION, ... | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
5FGR
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG - P21212 space group with Yb Heavy atom | | Descriptor: | Cell surface protein SpaA, YTTERBIUM (III) ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FIE
 
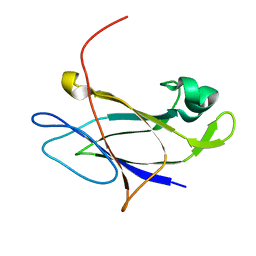 | | Crystal structure of N-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | Cell surface protein SpaA, SODIUM ION | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
2X9Z
 
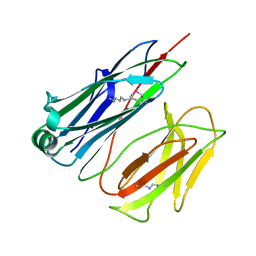 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Y
 
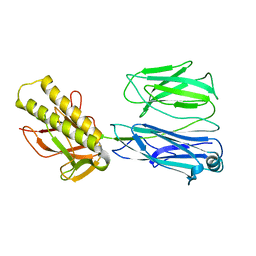 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9X
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
3HTL
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly with stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, Putative surface-anchored fimbrial subunit, SODIUM ION | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HR6
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, IODIDE ION, Putative surface-anchored fimbrial subunit | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7T31
 
 | | X-ray Structure of Clostridiodies difficile PilW | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Putative pilin protein chimera | | Authors: | Ronish, L.A, Piepenbrink, K.H. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
J.Biol.Chem., 298, 2022
|
|
5F44
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | ACETATE ION, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-03 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5FAA
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6JZJ
 
 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JZK
 
 | | Structure of FimA type-1 (FimA1) prepilin of the type V major fimbrium | | Descriptor: | GLYCEROL, Major fimbrium subunit FimA type-1, S,R MESO-TARTARIC ACID, ... | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
5AX6
 
 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | Descriptor: | ACETATE ION, CofB | | Authors: | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-03-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
2CO4
 
 | | Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide | | Descriptor: | PUTATIVE OUTER MEMBRANE PROTEIN | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
