5EFN
 
 | |
7THF
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B53 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)pentanedioic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
6TGD
 
 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | Descriptor: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6BXR
 
 | | Crystal structure of Toxoplasma gondii Mitochondrial Association Factor 1 B (MAF1B) | | Descriptor: | BROMIDE ION, GLYCEROL, Mitochondrial association factor 1, ... | | Authors: | Parker, M.L, Ramaswamy, R, Boulanger, M.J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Toxoplasma gondii locus required for the direct manipulation of host mitochondria has maintained multiple ancestral functions.
Mol. Microbiol., 108, 2018
|
|
7THD
 
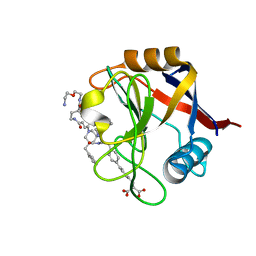 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B52 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, [(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)methyl]propanedioic acid | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
9NJF
 
 | |
4MU3
 
 | | The form A structure of an E21Q catalytic mutant of A. thaliana IGPD2 in complex with Mn2+ and a mixture of its substrate, 2R3S-IGP, and an inhibitor, 2S3S-IGP, to 1.12 A resolution | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, 1,2-ETHANEDIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
7TH7
 
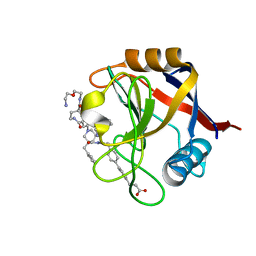 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B23 | | Descriptor: | 3-(4'-{[(4S,7S,11R,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,13,14,15,16,17,18,19,20,21,22-icosahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)propanoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
6BSX
 
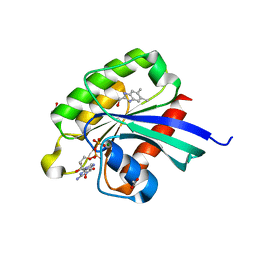 | |
6TD9
 
 | | X-ray structure of mature PA1624 from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PA1624, ... | | Authors: | Feiler, C.G, Blankenfeldt, W. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7THC
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B25 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)prop-2-ynoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
2QXH
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5V3D
 
 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae with bound fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FOSFOMYCIN, ... | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5EH5
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
7T7L
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
4N1I
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maidys | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
6MH5
 
 | |
2R1U
 
 | |
3KZU
 
 | |
5UNB
 
 | |
3KOQ
 
 | |
5EHN
 
 | | mAChE-syn TZ2PA5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[3-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
5EHR
 
 | |
5EJ0
 
 | | The vaccinia virus H3 envelope protein, a major target of neutralizing antibodies, exhibits a glycosyltransferase fold and binds UDP-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Gitti, R.K, Ostazesky, S.A, Su, H.P, Garboczi, D.N. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose.
J.Virol., 90, 2016
|
|
5AP3
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
