7LZF
 
 | | Crystal Structure of SETD2 bound to Compound 57 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-N-[(1R,3S)-3-{(3S)-3-[(methanesulfonyl)(methyl)amino]pyrrolidin-1-yl}cyclohexyl]-7-methyl-1H-indole-2-carboxamide, Histone-lysine N-methyltransferase SETD2, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
6HYI
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.4 A resolution in complex with inosine | | Descriptor: | INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.39944851 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
1O9A
 
 | | Solution structure of the complex of 1F12F1 from fibronectin with B3 from FnBB from S. dysgalactiae | | Descriptor: | FIBRONECTIN, FIBRONECTIN BINDING PROTEIN | | Authors: | Schwarz-Linek, U, Werner, J.M, Pickford, A.R, Pilka, E.S, Gurusiddappa, S, Briggs, J.A.G, Hook, M, Campbell, I.D, Potts, J.R. | | Deposit date: | 2002-12-11 | | Release date: | 2003-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper.
Nature, 423, 2003
|
|
3P16
 
 | | Crystal structure of DNA polymerase III sliding clamp | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Gui, W.J, Lin, S.Q, Chen, Y.Y, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2010-09-30 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of DNA polymerase III beta sliding clamp from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
1OIG
 
 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
5TUP
 
 | | X-ray Crystal Structure of the Aspergillus fumigatus Sliding Clamp | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Bruning, J.B, Marshall, A.C, Kroker, A.J, Wegener, K.L, Rajapaksha, H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with Human p21.
FEBS J., 284, 2017
|
|
2UXP
 
 | | TtgR in complex Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-29 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
8FA4
 
 | |
1QG9
 
 | | SECOND REPEAT (IS2MIC) FROM VOLTAGE-GATED SODIUM CHANNEL | | Descriptor: | PROTEIN (SODIUM CHANNEL PROTEIN, BRAIN II ALPHA SUBUNIT) | | Authors: | Doak, D.J, Mulvey, D, Kawaguchi, K, Villalain, J, Campbell, I.D. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies of synthetic peptides dissected from the voltage-gated sodium channel.
J.Mol.Biol., 258, 1996
|
|
6HSD
 
 | | Crystal structure of the oxidized form of the transcription regulator RsrR | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-09-30 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
6HSE
 
 | | Structure of dithionite-reduced RsrR in spacegroup P2(1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Rrf2 family transcriptional regulator, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
1SPQ
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
2O3Z
 
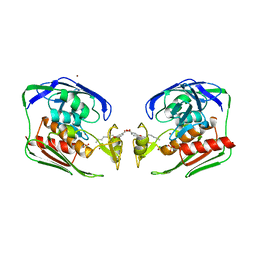 | | X-ray crystal structure of LpxC complexed with 3-heptyloxybenzoate | | Descriptor: | 3-(heptyloxy)benzoic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Christianson, D.W. | | Deposit date: | 2006-12-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Amphipathic benzoic acid derivatives: synthesis and binding in the hydrophobic tunnel of the zinc deacetylase LpxC.
Bioorg.Med.Chem., 15, 2007
|
|
6HSM
 
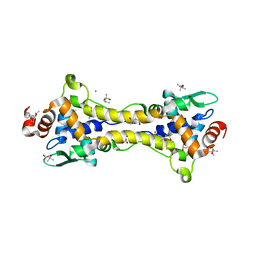 | | Structure of partially reduced RsrR in space group P2(1)2(1)2(1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
1PG9
 
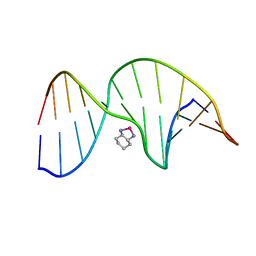 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
1SQD
 
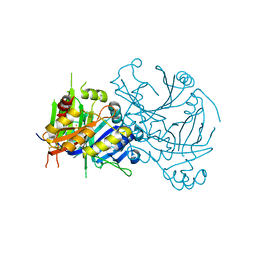 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1SQI
 
 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvic acid dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1SSG
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW0
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW3
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant T175V | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1TFZ
 
 | | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1SWU
 
 | | STREPTAVIDIN MUTANT Y43F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Atomic resolution structure of biotin-free Tyr43Phe streptavidin: what is in the binding site?
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3R90
 
 | | Crystal structure of Malignant T cell-amplified sequence 1 protein | | Descriptor: | GLYCEROL, Malignant T cell-amplified sequence 1, SULFATE ION, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Tong, Y, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Malignant T cell-amplified sequence 1 protein
to be published
|
|
