9HB0
 
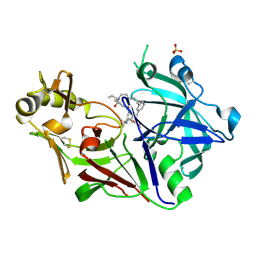 | | Crystal structure of Plasmodium falciparum Plasmepsin X in complex with the hydroxyethylamine drug 7k. | | Descriptor: | (4S)-4-[(1R)-2-[2-(3-methoxyphenyl)propan-2-ylamino]-1-oxidanyl-ethyl]-16-propyl-3,16-diazatricyclo[16.3.1.1^{6,10}]tricosa-1(21),6,8,10(23),18(22),19-hexaene-2,17-dione, Plasmepsin X, SULFATE ION | | Authors: | Withers-Martinez, C, George, R, Ogrodowicz, R, Kunzelmann, S, Purkiss, A, Kjaer, S, Walker, P, Kovada, V, Jirgensons, A, Blackman, M.J. | | Deposit date: | 2024-11-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Plasticity of Plasmodium falciparum Plasmepsin X to Accommodate Binding of Potent Macrocyclic Hydroxyethylamine Inhibitors.
J.Mol.Biol., 437, 2025
|
|
4ZWZ
 
 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX Using Structure-Activity Relationships of Glucosyl-Based Sulfamates.
J.Med.Chem., 58, 2015
|
|
9HG4
 
 | | NACHT domain of NLRP3 in complex with DFV890 | | Descriptor: | 1-[azanyl-oxidanylidene-[2-(2-oxidanylpropan-2-yl)-1,3-thiazol-5-yl]-$l^{6}-sulfanylidene]-3-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Dekker, C. | | Deposit date: | 2024-11-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.771 Å) | | Cite: | Discovery of DFV890, a Potent Sulfonimidamide-Containing NLRP3 Inflammasome Inhibitor.
J.Med.Chem., 68, 2025
|
|
7V7Q
 
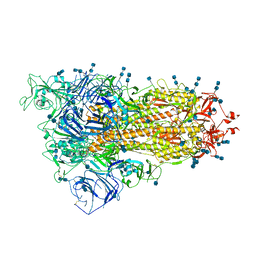 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 3
To Be Published
|
|
8ZHC
 
 | |
5DT2
 
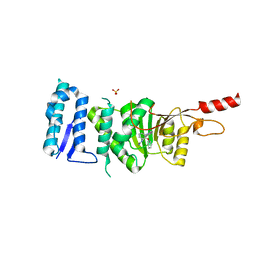 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
7V7P
 
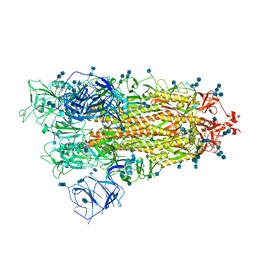 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 2
To Be Published
|
|
4ZXX
 
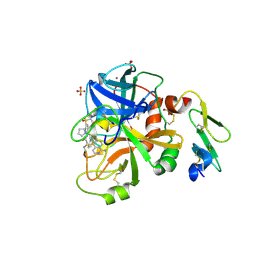 | |
3IL6
 
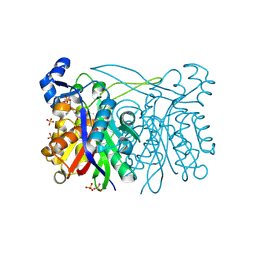 | | Structure of E. faecalis FabH in complex with 2-({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxybenzoyl}amino)benzoic acid | | Descriptor: | 2-[({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxyphenyl}carbonyl)amino]benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SULFATE ION | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
6TVR
 
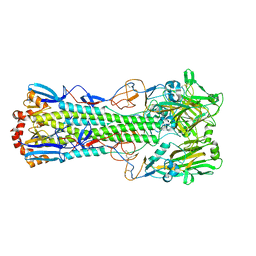 | | Crystal structure of the haemagglutinin mutant (Gln226Leu) from an H10N7 seal influenza virus isolated in Germany | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
7XE7
 
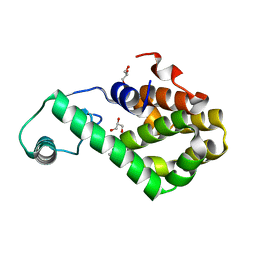 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH10 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
3IOQ
 
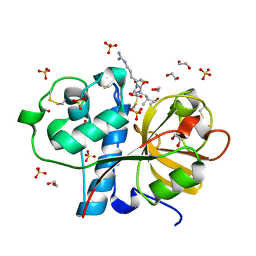 | | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64. | | Descriptor: | 1,2-ETHANEDIOL, CMS1MS2, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Gomes, M.T.R, Teixeira, R.D, Salas, C.E, Nagem, R.A.P. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64
To be Published
|
|
5E2O
 
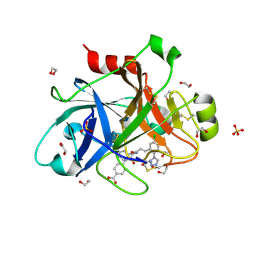 | |
6TVS
 
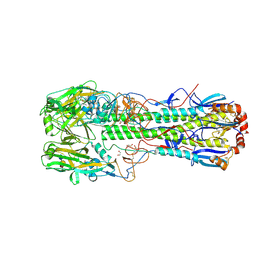 | | Crystal structure of the haemagglutinin mutant (Gln226Leu) from an H10N7 seal influenza virus isolated in Germany in complex with avian receptor analogue 3'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
4K8W
 
 | | An arm-swapped dimer of the S. pyogenes pilin specific assembly factor SipA | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, LepA | | Authors: | Young, P.G, Kang, H.J, Baker, E.N. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An arm-swapped dimer of the Streptococcus pyogenes pilin specific assembly factor SipA.
J.Struct.Biol., 183, 2013
|
|
7XE6
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
6QOG
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 10 (2-Amino-5-bromobenzothiazole) | | Descriptor: | 5-bromanyl-1,3-benzothiazol-2-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6EZ6
 
 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
7XE5
 
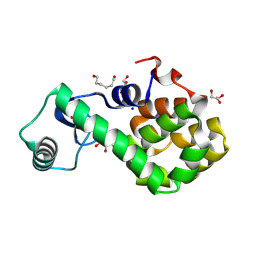 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
9INE
 
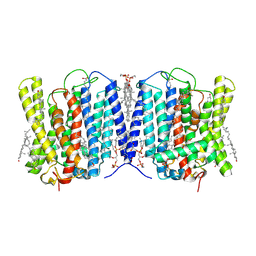 | | Cryo-EM structure of human XPR1 in closed state in the presence of KIDINS220-1-432 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ARACHIDONIC ACID, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9INH
 
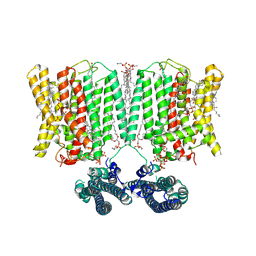 | | Cryo-EM structure of human XPR1 in complex with InsP6 in outward-facing state (SPX visible)-in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
5U2K
 
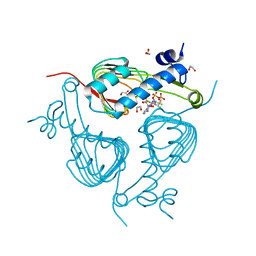 | | Crystal structure of Galactoside O-acetyltransferase complex with CoA (H3 space group) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Knapik, A.A, Niedzialkowska, E, Siuda, M.K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Galactoside O-acetyltransferase complex
with CoA (H3 space group)
to be published
|
|
6U4T
 
 | | Carbonic anhydrase 9 in complex with SB4197 | | Descriptor: | 4-methyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-08-26 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
6EVB
 
 | | Structure of E282Q A. niger Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
