2A3Y
 
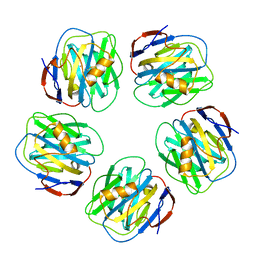 | | Pentameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy-2-methyl-1,3-dioxane]-5-yloxycarbamoyl}-ethane. | | Descriptor: | BIS-1,2-{[(Z)-2-CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBAMOYL}-ETHANE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
4R8H
 
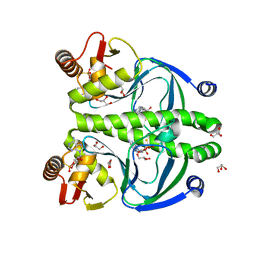 | | The role of protein-ligand contacts in allosteric regulation of the Escherichia coli Catabolite Activator Protein | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, GLYCEROL, cAMP-activated global transcriptional regulator CRP | | Authors: | Townsend, P.D, Pohl, E, McLeish, T.C.B, Rodgers, T.L, Glover, L.C, Korhonen, H.J, Wilson, M.R, Hodgson, D.R.W, Cann, M.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein.
J.Biol.Chem., 290, 2015
|
|
3GMQ
 
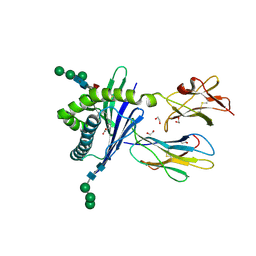 | | Structure of mouse CD1d expressed in SF9 cells, no ligand added | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
223L
 
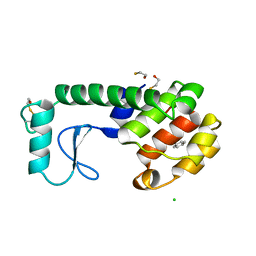 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
5KCE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCD
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
7A9I
 
 | |
7A9J
 
 | |
1QKU
 
 | | WILD TYPE ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism.
J. Biol. Chem., 276, 2001
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
6G4Q
 
 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
2JGC
 
 | | Structure of the human eIF4E homologous protein, 4EHP without ligand bound | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
8EVZ
 
 | | DdlB from Pseudomonas aeruginosa PAO1 in complex with ADP and phosphorylated D-cycloserine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase B, MAGNESIUM ION, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
8EVW
 
 | | DdlA from Pseudomonas aeruginosa PAO1 in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase A, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
8EVX
 
 | | DdlA from Pseudomonas aeruginosa PAO1 in complex with ADP and phosphorylated D-cycloserine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase A, MAGNESIUM ION, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
8EVY
 
 | | DdlB from Pseudomonas aeruginosa PAO1 in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase B, ... | | Authors: | Pederick, J.L, Woolman, J.C, Bruning, J.B. | | Deposit date: | 2022-10-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparative functional and structural analysis of Pseudomonas aeruginosa d-alanine-d-alanine ligase isoforms as prospective antibiotic targets.
Febs J., 290, 2023
|
|
9FSK
 
 | | Crystal structure of the HECT domain of Smurf1 | | Descriptor: | E3 ubiquitin-protein ligase SMURF1 | | Authors: | Ostermann, N. | | Deposit date: | 2024-06-21 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Therapeutic potential of allosteric HECT E3 ligase inhibition.
Cell, 188, 2025
|
|
3B76
 
 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
4KCD
 
 | | Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State | | Descriptor: | GLYCEROL, Glutamate receptor ionotropic, NMDA 3A | | Authors: | Yao, Y, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
