6ICD
 
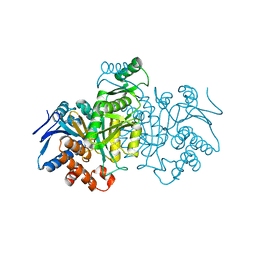 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
5ZT1
 
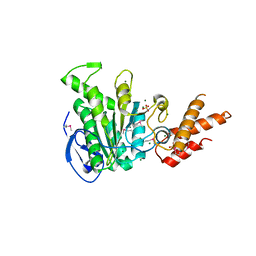 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
1GS4
 
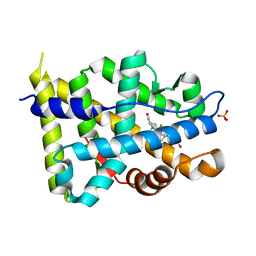 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3OKL
 
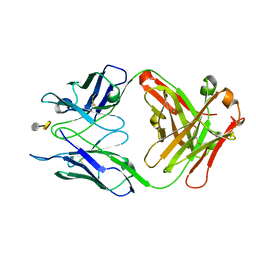 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2ZBG
 
 | | Calcium pump crystal structure with bound AlF4 and TG in the absence of calcium | | Descriptor: | MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7LNU
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNW
 
 | |
7LNT
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to benzyl monophosphate and ATP | | Descriptor: | (phenylmethyl) dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
5YGX
 
 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
7LNV
 
 | | Apo Structure of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus | | Descriptor: | GLYCEROL, Isopentenyl phosphate kinase, MALONATE ION | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
6SW5
 
 | | Crystal structure of the human S-adenosylmethionine synthetase 1 (ligand-free form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, S-adenosylmethionine synthase isoform type-1 | | Authors: | Panmanee, J, Antoyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the dominant inheritance of hypermethioninemia associated with the Arg264His mutation in the MAT1A gene.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4QEI
 
 | | Two distinct conformational states of GlyRS captured in crystal lattice | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine--tRNA ligase, tRNA-Gly-CCC-2-2 | | Authors: | Xie, W, Qin, X, Deng, X, Zhang, Q, Li, Q. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
3OKO
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
5YLX
 
 | | Integrated illustration of a valid epitope based on the SLA class I structure and tetramer technique could carry forward the development of molecular vaccine in swine species | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PRRSV-NSP9-TMP9 peptide | | Authors: | Pan, X.C, Wei, X.H, Zhang, N, Xia, C. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I).
Front Immunol, 10, 2019
|
|
8SLY
 
 | | Rat TRPV2 bound with 2 CBD ligands in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
2HZ7
 
 | | Crystal structure of the Glutaminyl-tRNA synthetase from Deinococcus radiodurans | | Descriptor: | Glutaminyl-tRNA synthetase | | Authors: | Deniziak, M, Sauter, C, Becker, H.D, Paulus, C, Giege, R, Kern, D. | | Deposit date: | 2006-08-08 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation
Nucleic Acids Res., 35, 2007
|
|
7QAY
 
 | |
2VDK
 
 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
7QAT
 
 | |
7QAW
 
 | |
7QAX
 
 | |
1NS6
 
 | |
7QAU
 
 | |
7QAQ
 
 | |
