4RRR
 
 | |
5T74
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-(4-sulfamoylphenyl)ethanamide, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
8E4O
 
 | | The closed C1-state mouse TRPM8 structure in complex with putative PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4N
 
 | | The closed C1-state mouse TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4M
 
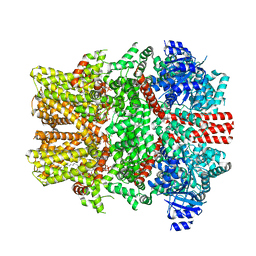 | | The intermediate C2-state mouse TRPM8 structure in complex with the cooling agonist C3 and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E9P
 
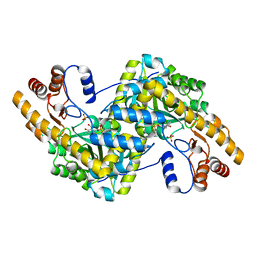 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9J
 
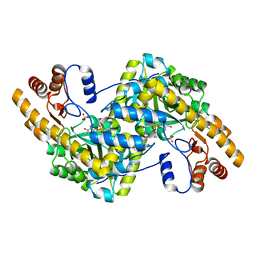 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9U
 
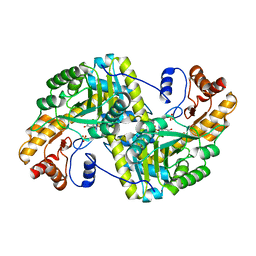 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
3V80
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-O-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-O-prop-2-yn-1-yladenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
7MZ5
 
 | |
3V7W
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-azido-5'-deoxyadenosine | | Descriptor: | 5'-azido-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.0102 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
1RH3
 
 | |
2FPI
 
 | |
3V8M
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with 5'-azido-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8Q
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-5'-deoxyadenosine | | Descriptor: | 5'-amino-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3GI6
 
 | | Crystal structure of protease inhibitor, AD78 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2R)-2-Hydroxy-3-[[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino]-1-(phenylmethyl)propyl]-2-oxo-3-[3-(trif luoromethyl)phenyl]-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
8I1Y
 
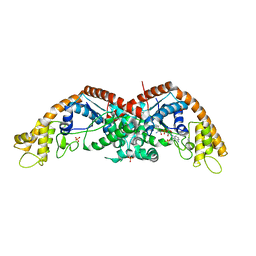 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
3G5Y
 
 | |
8I27
 
 | |
8I2A
 
 | |
4RRK
 
 | |
8I1W
 
 | |
8I2L
 
 | |
8I2C
 
 | |
