1VD6
 
 | |
1VD7
 
 | | Solution structure of FMBP-1 tandem repeat 1 | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 1
To be Published
|
|
1VD8
 
 | | Solution structure of FMBP-1 tandem repeat 2 | | Descriptor: | fibroin-modulator-binding-protein-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 2
To be Published
|
|
1VD9
 
 | | Solution structure of FMBP-1 tandem repeat 3 | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 3
To be Published
|
|
1VDA
 
 | | Solution structure of FMBP-1 tandem repeat 4 | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 4
To be Published
|
|
1VDB
 
 | | NMR structure of FMBP-1 tandem repeat 1 in 30%(v/v) TFE solution | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of FMBP-1 tandem repeat 1 in 30%(v/v) TFE solution
To be Published
|
|
1VDC
 
 | | STRUCTURE OF NADPH DEPENDENT THIOREDOXIN REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DEPENDENT THIOREDOXIN REDUCTASE, SULFATE ION | | Authors: | Dai, S, Eklund, H. | | Deposit date: | 1996-09-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 A resolution.
J.Mol.Biol., 264, 1996
|
|
1VDD
 
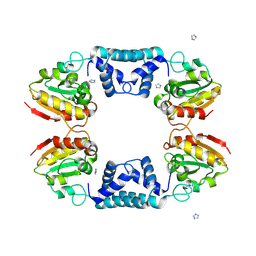 | |
1VDE
 
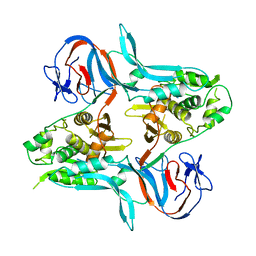 | |
1VDF
 
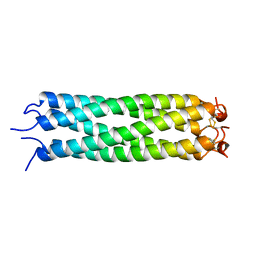 | |
1VDG
 
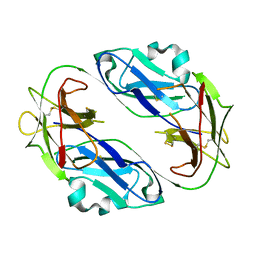 | | Crystal structure of LIR1.01, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2004-03-22 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of LIR1.03, one of the alleles of LIR1
To be Published
|
|
1VDH
 
 | | Structure-based functional identification of a novel heme-binding protein from thermus thermophilus HB8 | | Descriptor: | muconolactone isomerase-like protein | | Authors: | Ebihara, A, Okamoto, A, Kousumi, Y, Yamamoto, H, Masui, R, Ueyama, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1VDI
 
 | | Solution structure of actin-binding domain of troponin in Ca2+-free state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDJ
 
 | | Solution structure of actin-binding domain of troponin in Ca2+-bound state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDK
 
 | |
1VDL
 
 | | Solution Structure of RSGI RUH-013, a UBA domain in Mouse cDNA | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Doi-Katayama, Y, Hirota, H, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-23 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-013, a UBA domain in Mouse cDNA
To be Published
|
|
1VDM
 
 | |
1VDN
 
 | |
1VDP
 
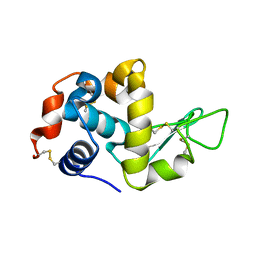 | | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space
to be published
|
|
1VDQ
 
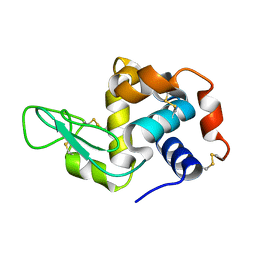 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
1VDR
 
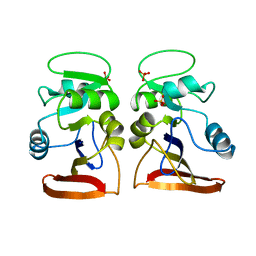 | | DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, PHOSPHATE ION | | Authors: | Pieper, U, Herzberg, O. | | Deposit date: | 1997-11-30 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
Structure, 6, 1998
|
|
1VDS
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
1VDT
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
1VDV
 
 | | Bovine Milk Xanthine Dehydrogenase Y-700 Bound Form | | Descriptor: | 1-[3-CYANO-4-(NEOPENTYLOXY)PHENYL]-1H-PYRAZOLE-4-CARBOXYLIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Fukunari, A, Okamoto, K, Nishino, T, Eger, B.T, Pai, E.F, Kamezawa, M, Yamada, I, Kato, N. | | Deposit date: | 2004-03-25 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Y-700 [1-[3-Cyano-4-(2,2-dimethylpropoxy)phenyl]-1H-pyrazole-4-carboxylic acid]: a potent xanthine oxidoreductase inhibitor with hepatic excretion
J.Pharmacol.Exp.Ther., 311, 2004
|
|
1VDW
 
 | |
