8CKP
 
 | | X-ray structure of the crystallization-prone form of subfamily III haloalkane dehalogenase DhmeA from Haloferax mediterranei | | Descriptor: | Alpha/beta fold hydrolase, CHLORIDE ION | | Authors: | Marek, M, Chmelova, K, Schenkmayerova, A, Croll, T, Read, R.J, Diederichs, K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Multimeric structure of a subfamily III haloalkane dehalogenase-like enzyme solved by combination of cryo-EM and x-ray crystallography.
Protein Sci., 32, 2023
|
|
5LIP
 
 | | PSEUDOMONAS LIPASE COMPLEXED WITH RC-(RP, SP)-1,2-DIOCTYLCARBAMOYLGLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER, TRIACYL-GLYCEROL HYDROLASE | | Authors: | Lang, D.A, Dijkstra, B.W. | | Deposit date: | 1997-09-02 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the chiral selectivity of Pseudomonas cepacia lipase
Eur.J.Biochem., 254, 1998
|
|
6NY9
 
 | | Alpha/beta hydrolase domain-containing protein 10 from mouse | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycophenolic acid acyl-glucuronide esterase, mitochondrial, ... | | Authors: | Cao, Y, Rice, P.A, Dickinson, B.C. | | Deposit date: | 2019-02-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5.
Nat.Chem.Biol., 15, 2019
|
|
5O2I
 
 | | An efficient setup for fixed-target, time-resolved serial crystallography with optical excitation | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mueller-Werkmeister, H, Mehrabi, P, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2017-05-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An efficient setup for fixed-target, time-resolved serial crystallography with optical excitation
To Be Published
|
|
6PUX
 
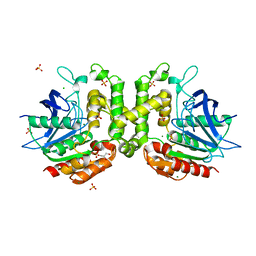 | |
6TY7
 
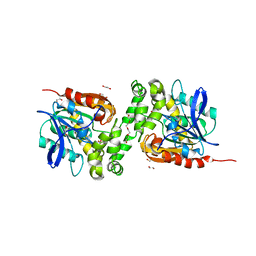 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
7TVW
 
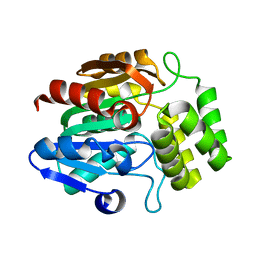 | | Crystal structure of Arabidopsis thaliana DLK2 | | Descriptor: | Alpha/beta-Hydrolases superfamily protein | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
7UOC
 
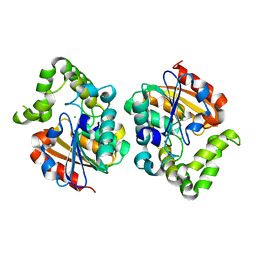 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
7SFM
 
 | | Mycobacterium tuberculosis Hip1 crystal structure | | Descriptor: | ACETATE ION, GLYCEROL, Hip1 | | Authors: | Ostrov, D.A, Li, D. | | Deposit date: | 2021-10-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
6V7N
 
 | |
7RYT
 
 | |
5EGN
 
 | |
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
3KXP
 
 | | Crystal Structure of E-2-(Acetamidomethylene)succinate Hydrolase | | Descriptor: | Alpha-(N-acetylaminomethylene)succinic acid hydrolase, CHLORIDE ION | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure determination and characterization of the vitamin B(6) degradative enzyme (E)-2-(acetamidomethylene)succinate hydrolase.
Biochemistry, 49, 2010
|
|
5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
5MXP
 
 | |
3LIP
 
 | |
5FLK
 
 | |
5NYV
 
 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein crystals IR laser ablated from aqueous solution at high speed retain their diffractive properties: applications in high-speed serial crystallography.
J.Appl.Crystallogr., 50, 2017
|
|
5O2G
 
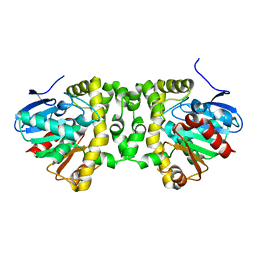 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | Deposit date: | 2017-05-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography
To Be Published
|
|
5HZ2
 
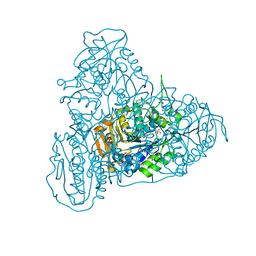 | | Crystal structure of PhaC1 from Ralstonia eutropha | | Descriptor: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | Authors: | Kim, J, Kim, K.-J. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-07 | | Last modified: | 2017-04-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
1K63
 
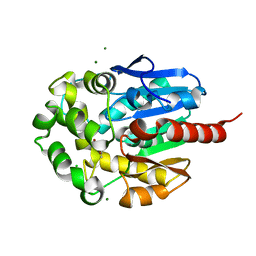 | | Complex of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis with UT26 2-BROMO-2-PROPENE-1-OL at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, 2-BROMO-2-PROPENE-1-OL, BROMIDE ION, ... | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1K6E
 
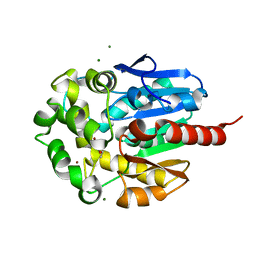 | | COMPLEX OF HYDROLYTIC HALOALKANE DEHALOGENASE LINB FROM SPHINGOMONAS PAUCIMOBILIS UT26 WITH 1,2-PROPANEDIOL (PRODUCT OF DEHALOGENATION OF 1,2-DIBROMOPROPANE) AT 1.85A | | Descriptor: | 1-BROMOPROPANE-2-OL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Streltsov, V.A, Prokop, Z, Damborsky, J, Nagata, Y, Oakley, A, Wilce, M.C.J. | | Deposit date: | 2001-10-16 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates.
Biochemistry, 42, 2003
|
|
6T6H
 
 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6T6Z
 
 | |
