6Q45
 
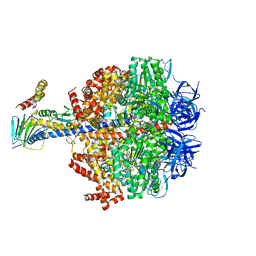 | | F1-ATPase from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Petri, J, Nakatani, Y, Montgomery, M.G, Ferguson, S.A, Aragao, D, Leslie, A.G.W, Heikal, A, Walker, J.E, Cook, G.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of F1-ATPase from the obligate anaerobe Fusobacterium nucleatum.
Open Biology, 9, 2019
|
|
1MAB
 
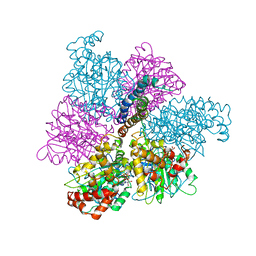 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2XND
 
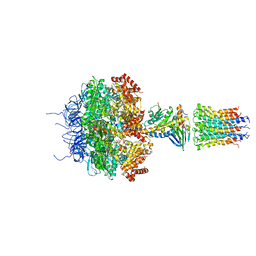 | | Crystal structure of bovine F1-c8 sub-complex of ATP Synthase | | Descriptor: | ATP SYNTHASE LIPID-BINDING PROTEIN, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Watt, I.N, Montgomery, M.G, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8SPW
 
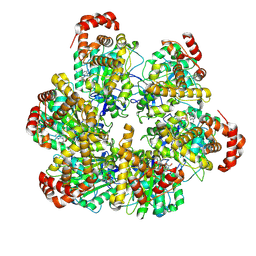 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
4ASU
 
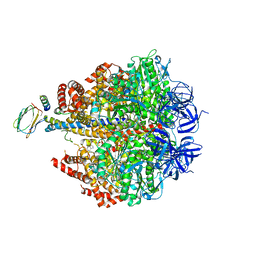 | | F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Rees, D.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2012-05-03 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence of a New Catalytic Intermediate in the Pathway of ATP Hydrolysis by F1-ATPase from Bovine Heart Mitochondria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8KI3
 
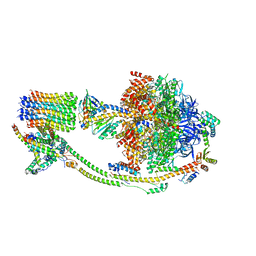 | |
3FKS
 
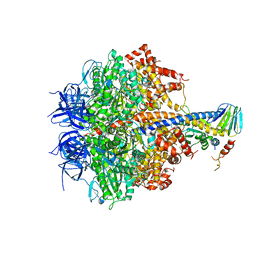 | | Yeast F1 ATPase in the absence of bound nucleotides | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Kabaleeswaran, V, Symersky, J, Shen, H, Walker, J.E, Leslie, A.G.W, Mueller, D.M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.587 Å) | | Cite: | Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
J.Biol.Chem., 284, 2009
|
|
1KMH
 
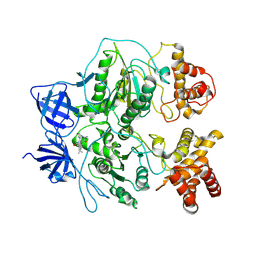 | |
1SKY
 
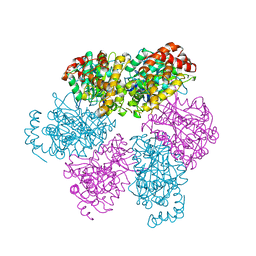 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
8SPV
 
 | | PS3 F1 Rotorless, no ATP | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8SPX
 
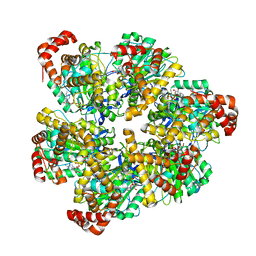 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
5IK2
 
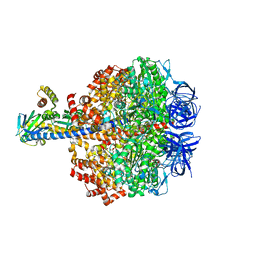 | | Caldalaklibacillus thermarum F1-ATPase (epsilon mutant) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XD7
 
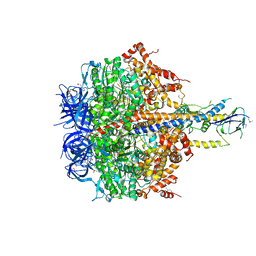 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1W0K
 
 | | ADP inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
1W0J
 
 | | Beryllium fluoride inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
2CK3
 
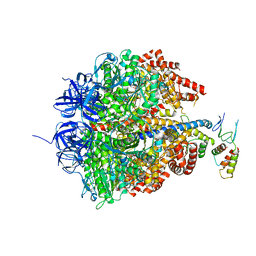 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5HKK
 
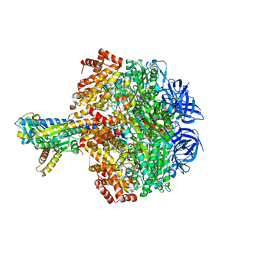 | | Caldalaklibacillus thermarum F1-ATPase (wild type) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YXW
 
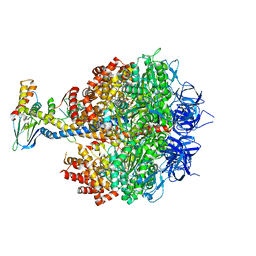 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1COW
 
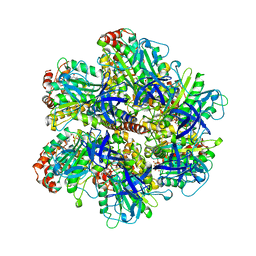 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2F43
 
 | | Rat liver F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase alpha chain, ... | | Authors: | Chen, C, Saxena, A.K, Simcoke, W.N, Garboczi, D.N, Pedersen, P.L, Ko, Y.H. | | Deposit date: | 2005-11-22 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism.
J.Biol.Chem., 281, 2006
|
|
4TT3
 
 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TSF
 
 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
7P3W
 
 | | F1Fo-ATP synthase from Acinetobacter baumannii (state 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Demmer, J.K, Phillips, B.P, Uhrig, O.L, Filloux, A, Allsopp, L.P, Bublitz, M, Meier, T. | | Deposit date: | 2021-07-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of ATP synthase from ESKAPE pathogen Acinetobacter baumannii.
Sci Adv, 8, 2022
|
|
