9EQ3
 
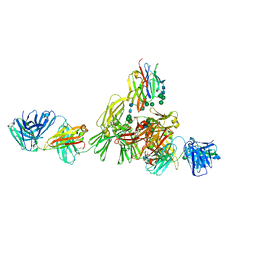 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The dynamics of hinge flexibility in receptor bound immunoglobulin E revealed by electron microscopy
Biorxiv, 2023
|
|
9EQ4
 
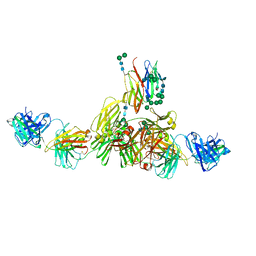 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The dynamics of hinge flexibility in receptor bound immunoglobulin E revealed by electron microscopy
Biorxiv, 2023
|
|
8R61
 
 | |
