1Q14
 
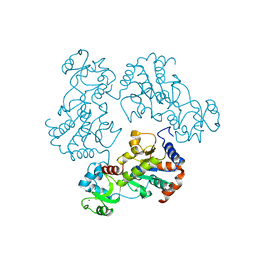 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | Descriptor: | CHLORIDE ION, HST2 protein, ZINC ION | | Authors: | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SZC
 
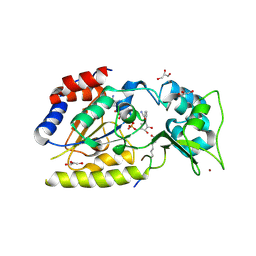 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q17
 
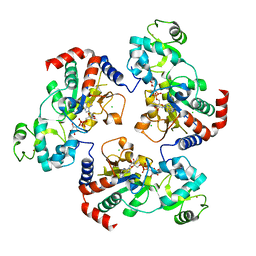 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, HST2 protein, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2'-O-Acetyl
ADP Ribose and Histone Peptide.
Structure, 11, 2003
|
|
1Q1A
 
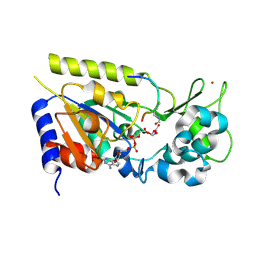 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, HST2 protein, Histone H4, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-Acetyl ADP ribose and histone peptide.
Structure, 11, 2003
|
|
