8R19
 
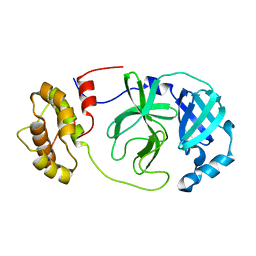 | | SARS-CoV-2 Mpro (Omicron, P132H) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R0V
 
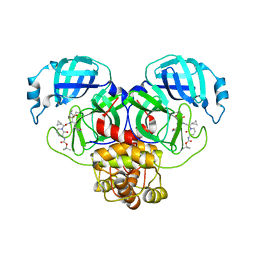 | | SARS-CoV-2 Mpro (Omicron, P132H) in complex with alpha-ketoamide 13b-K at pH 6.5 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R26
 
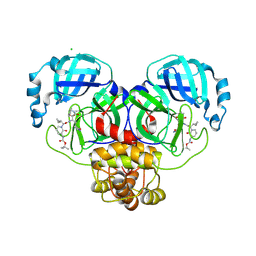 | | SARS-CoV-2 Mpro (Omicron,P132H) in complex with alpha-ketoamide 13b-K at pH 8.5 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-03 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R1Q
 
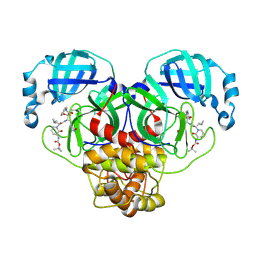 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) in complex with alpha-ketoamide 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Sun, X, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R24
 
 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Sun, X, Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
