1T9T
 
 | |
1T9U
 
 | |
1T9V
 
 | |
1T9W
 
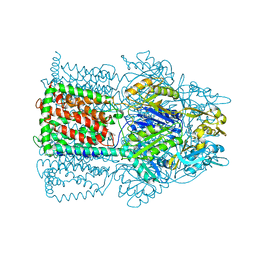
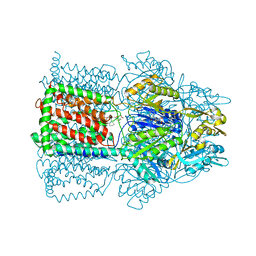 | | Structural Basis of Multidrug Transport by the AcrB Multidrug Efflux Pump | | Descriptor: | 6-[[(2-ETHOXY-1-NAPHTHALENYL)CARBONYL]AMINO]-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Nikaido, H. | | Deposit date: | 2004-05-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | A Periplasmic Drug-Binding Site of the AcrB Multidrug Efflux Pump: a Crystallographic and Site-Directed Mutagenesis Study
J.Bacteriol., 187, 2005
|
|
1T9X
 
 | |
1T9Y
 
 | |
1T9Z
 
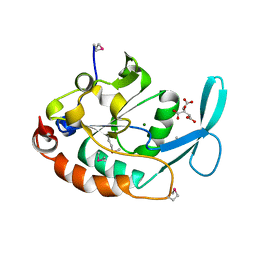 | | Three-dimensional structure of a RNA-polymerase II binding protein. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of
RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
1TA0
 
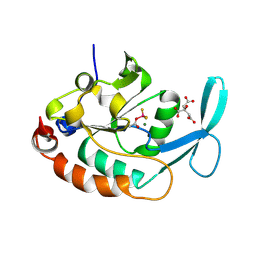 | | Three-dimensional structure of a RNA-polymerase II binding protein with associated ligand. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, T, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
1TA1
 
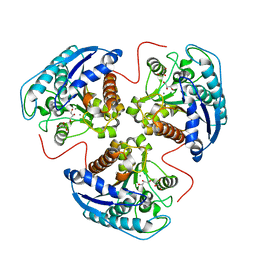 | | H141C mutant of rat liver arginase I | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-19 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1TA2
 
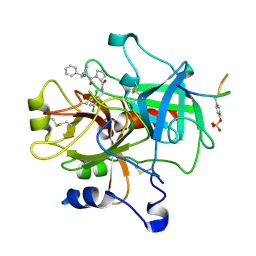 | | Crystal structure of thrombin in complex with compound 1 | | Descriptor: | 1-(2-AMINO-3,3-DIPHENYL-PROPIONYL)-PYRROLIDINE-3-CARBOXYLIC ACID 2,5-DICHLORO-BENZYLAMIDE, Hirudin, thrombin | | Authors: | Tucker, T.J, Brady, S.F, Lumma, W.C, Lewis, S.D, Gardel, S.J, Naylor-Olsen, A.M, Yan, Y, Sisko, J.T, Stauffer, K.J, Lucas, B.Y, Lynch, J.J, Cook, J.J, Stranieri, M.T, Holahan, M.A, Lyle, E.A, Baskin, E.P, Chen, I.-W, Dancheck, K.B, Krueger, J.A, Cooper, C.M, Vacca, J.P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of potent and orally bioavailable
noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position
J.Med.Chem., 41, 1998
|
|
1TA3
 
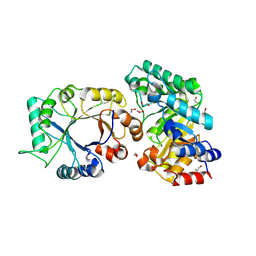 | | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
1TA4
 
 | |
1TA6
 
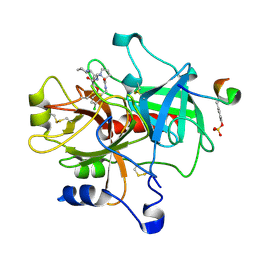 | | Crystal structure of thrombin in complex with compound 14b | | Descriptor: | 1-[2-AMINO-2-CYCLOHEXYL-ACETYL]-PYRROLIDINE-3-CARBOXYLIC ACID 5-CHLORO-2-(2-ETHYLCARBAMOYL-ETHOXY)-BENZYLAMIDE, Hirudin, thrombin | | Authors: | Tucker, T.J, Brady, S.F, Lumma, W.C, Lewis, S.D, Gardel, S.J, Naylor-Olsen, A.M, Yan, Y, Sisko, J.T, Stauffer, K.J, Lucas, B.Y, Lynch, J.J, Cook, J.J, Stranieri, M.T, Holahan, M.A, Lyle, E.A, Baskin, E.P, Chen, I.-W, Dancheck, K.B, Krueger, J.A, Cooper, C.M, Vacca, J.P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of a series of potent and orally bioavailable
noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position
J.Med.Chem., 41, 1998
|
|
1TA8
 
 | |
1TA9
 
 | |
1TAB
 
 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
1TAC
 
 | |
1TAD
 
 | | GTPASE MECHANISM OF GPROTEINS FROM THE 1.7-ANGSTROM CRYSTAL STRUCTURE OF TRANSDUCIN ALPHA-GDP-ALF4- | | Descriptor: | CACODYLATE ION, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sondek, J, Lambright, D.G, Noel, J.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1995-01-05 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4.
Nature, 372, 1994
|
|
1TAE
 
 | |
1TAF
 
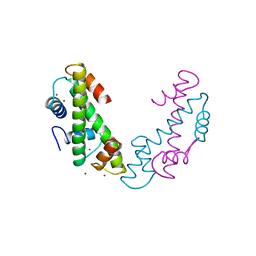 | | DROSOPHILA TBP ASSOCIATED FACTORS DTAFII42/DTAFII62 HETEROTETRAMER | | Descriptor: | TFIID TBP ASSOCIATED FACTOR 42, TFIID TBP ASSOCIATED FACTOR 62, ZINC ION | | Authors: | Xie, X, Kokubo, T, Cohen, S.L, Mirza, U.A, Hoffmann, A, Chait, B.T, Roeder, R.G, Nakatani, Y, Burley, S.K. | | Deposit date: | 1996-06-01 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural similarity between TAFs and the heterotetrameric core of the histone octamer.
Nature, 380, 1996
|
|
1TAG
 
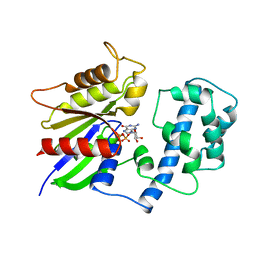 | | STRUCTURAL DETERMINANTS FOR ACTIVATION OF THE ALPHA-SUBUNIT OF A HETEROTRIMERIC G PROTEIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRANSDUCIN-ALPHA | | Authors: | Lambright, D.G, Noel, J.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein.
Nature, 369, 1994
|
|
1TAH
 
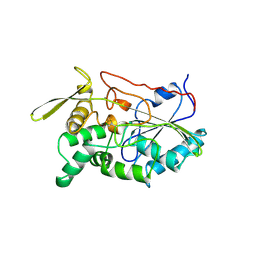 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
1TAL
 
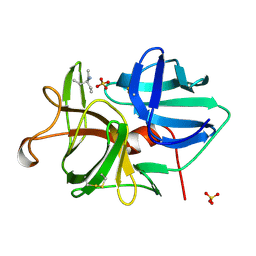 | | ALPHA-LYTIC PROTEASE AT 120 K (SINGLE STRUCTURE MODEL) | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Rader, S.D, Agard, D.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational substates in enzyme mechanism: the 120 K structure of alpha-lytic protease at 1.5 A resolution.
Protein Sci., 6, 1997
|
|
1TAM
 
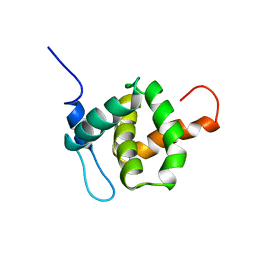 | | HUMAN IMMUNODEFICIENCY VIRUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 MATRIX PROTEIN | | Authors: | Matthews, S, Barlow, P, Clark, N, Kingsman, S, Kingsman, A, Campbell, I. | | Deposit date: | 1996-02-07 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of p17, the HIV matrix protein.
Biochem.Soc.Trans., 23, 1995
|
|
1TAN
 
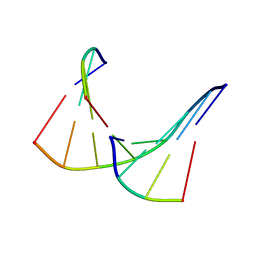 | | TANDEM DNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(P*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*AP*GP*CP*TP*G)-3') | | Authors: | Denisov, A, Sandstrom, A, Maltseva, T, Pyshnyi, D, Ivanova, E, Zarytova, V, Chattopadhyaya, J. | | Deposit date: | 1997-06-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of estrone (Es)-tethered tandem DNA duplex: [d(5'pCAGCp3')-Es] + [Es-d(5'pTCCA3')]: d(5'pTGGAGCTG3').
J.Biomol.Struct.Dyn., 15, 1997
|
|
