1OGM
 
 | |
1OGO
 
 | | Dex49A from Penicillium minioluteum complex with isomaltose | | Descriptor: | DEXTRANASE, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Larsson, A.M, Stahlberg, J, Jones, T.A. | | Deposit date: | 2003-05-08 | | Release date: | 2003-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dextranase from Penicillium Minioluteum. Reaction Course, Crystal Structure, and Product Complex
Structure, 11, 2003
|
|
1OGP
 
 | | The crystal structure of plant sulfite oxidase provides insight into sulfite oxidation in plants and animals | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(VI), CESIUM ION, GLYCEROL, ... | | Authors: | Schrader, N, Fischer, K, Theis, K, Mendel, R.R, Schwarz, G, Kisker, C. | | Deposit date: | 2003-05-08 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Plant Sulfite Oxidase Provides Insights Into Sulfite Oxidation in Plants and Animals
Structure, 11, 2003
|
|
1OGQ
 
 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OGS
 
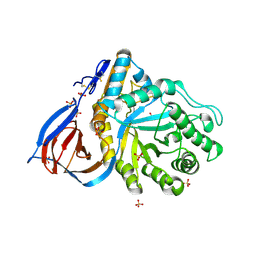 | | human acid-beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Dvir, H, Harel, M, McCarthy, A.A, Toker, L, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structure of Human Acid-Beta-Glucosidase, the Defective Enzyme in Gaucher Disease
Embo Rep., 4, 2003
|
|
1OGT
 
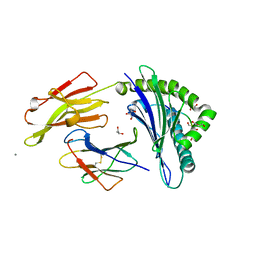 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE VASOACTIVE INTESTINAL PEPTIDE TYPE 1 RECEPTOR (VIPR) PEPTIDE (RESIDUES 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-05-13 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
J. Exp. Med., 199, 2004
|
|
1OGU
 
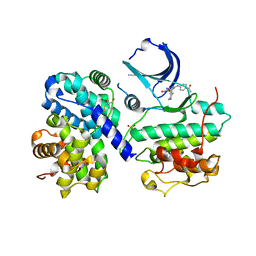 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2/CYCLIN A COMPLEXED WITH A 2-ARYLAMINO-4-CYCLOHEXYLMETHYL-5-NITROSO-6-AMINOPYRIMIDINE INHIBITOR | | Descriptor: | 4-{[4-AMINO-6-(CYCLOHEXYLMETHOXY)-5-NITROSOPYRIMIDIN-2-YL]AMINO}BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-05-13 | | Release date: | 2003-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1OGV
 
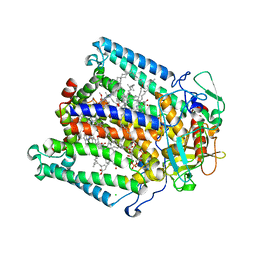 | | Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Katona, G, Andreasson, U, Landau, E.M, Andreasson, L.-E, Neutze, R. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lipidic Cubic Phase Crystal Structure of the Photosynthetic Reaction Centre from Rhodobacter Sphaeroides at 2.35 A Resolution
J.Mol.Biol., 331, 2003
|
|
1OGW
 
 | | Synthetic Ubiquitin with fluoro-Leu at 50 and 67 | | Descriptor: | UBIQUITIN | | Authors: | Alexeev, D, Ramage, R, Young, D.W, Sawyer, L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Synthesis, Structural and Biological Studies of Ubiquitin Mutants Containing (2S, 4S)-5-Fluoroleucine Residues Strategically Placed in the Hydrophobic Core
Chembiochem, 4, 2003
|
|
1OGX
 
 | | High Resolution Crystal Structure Of Ketosteroid Isomerase Mutant D40N(D38N, Ti Numbering) from Pseudomonas putida Complexed With Equilenin At 2.0 A Resolution. | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-05-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbation of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
1OGZ
 
 | | Crystal Structure Of 5-3-Ketosteroid Isomerase Mutants P39A Complexed With Equilenin From Pseudomonas Testosteroni | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Nam, G.H, Cha, S.-S, Yun, Y.S, Oh, Y.H, Hong, B.H, Lee, H.-S, Choi, K.Y. | | Deposit date: | 2003-05-20 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Conserved Cis-Pro39 Residue Plays a Crucial Role in the Proper Positioning of the Catalytic Base Asp38 in Ketosteroid Isomerase from Comamonas Testosteroni.
Biochem.J., 375, 2003
|
|
1OH0
 
 | | CRYSTAL STRUCTURE OF KETOSTEROID ISOMERASE COMPLEXED WITH EQUILENIN | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Kim, K.-H, Cha, S.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-06-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structures of Delta5-3-Ketosteroid Isomerase with and without a Reaction Intermediate Analogue
Biochemistry, 36, 1997
|
|
1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
1OH2
 
 | |
1OH3
 
 | | E78R mutant of a carbohydrate binding module family 29 | | Descriptor: | NON-CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nurizzo, D, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-Mediated Dimerization of a Carbohydrate -Binding Module Reveals a Novel Mechanism for Protein-Carbohydrate Recognition
J.Mol.Biol., 337, 2004
|
|
1OH4
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1OH5
 
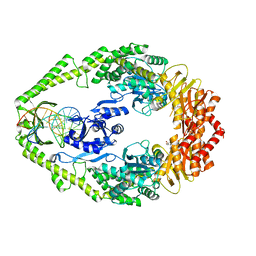 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
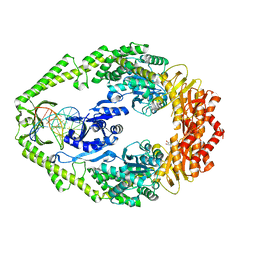 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH7
 
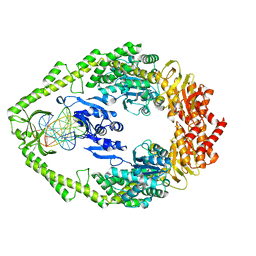 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A G:G MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*GP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH8
 
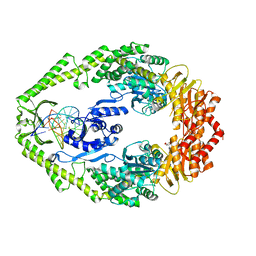 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN UNPAIRED THYMIDINE | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP* TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*CP*TP*TP*GP*GP*CP* AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH9
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP, N-acetyl-L-glutamate and the transition-state mimic AlF4- | | Descriptor: | ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OHA
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP and N-acetyl-L-glutamate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OHB
 
 | | Acetylglutamate kinase from Escherichia coli complexed with ADP and sulphate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OHC
 
 | |
