1ATK
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH THE COVALENT INHIBITOR E-64 | | Descriptor: | CATHEPSIN K, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1996-12-19 | | Release date: | 1998-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human osteoclast cathepsin K complex with E-64.
Nat.Struct.Biol., 4, 1997
|
|
1ATL
 
 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | Descriptor: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1ATN
 
 | | Atomic structure of the actin:DNASE I complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kabsch, W, Mannherz, H.G, Suck, D, Pai, E, Holmes, K.C. | | Deposit date: | 1991-03-08 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the actin:DNase I complex.
Nature, 347, 1990
|
|
1ATO
 
 | | THE STRUCTURE OF THE ISOLATED, CENTRAL HAIRPIN OF THE HDV ANTIGENOMIC RIBOZYME, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*CP*UP*CP*CP*UP*CP*GP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Kolk, M.H, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1997-08-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
EMBO J., 16, 1997
|
|
1ATP
 
 | | 2.2 angstrom refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MNATP and a peptide inhibitor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PEPTIDE INHIBITOR PKI(5-24), ... | | Authors: | Zheng, J, Trafny, E.A, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Teneyck, L.F, Sowadski, J.M. | | Deposit date: | 1993-01-08 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1ATR
 
 | |
1ATS
 
 | |
1ATT
 
 | |
1ATU
 
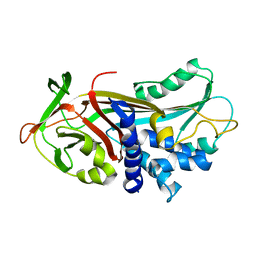 | | UNCLEAVED ALPHA-1-ANTITRYPSIN | | Descriptor: | ALPHA-1-ANTITRYPSIN | | Authors: | Ryu, S.-E, Choi, H.-J. | | Deposit date: | 1997-05-11 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The native strains in the hydrophobic core and flexible reactive loop of a serine protease inhibitor: crystal structure of an uncleaved alpha1-antitrypsin at 2.7 A.
Structure, 4, 1996
|
|
1ATV
 
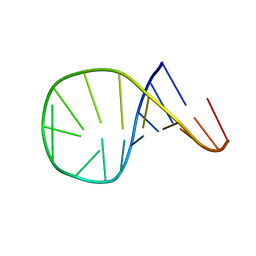 | | HAIRPIN WITH AGAA TETRALOOP, NMR, 4 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*AP*CP*CP*AP*GP*AP*AP*GP*GP*UP*CP*CP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1ATW
 
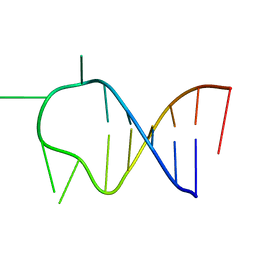 | | HAIRPIN WITH AGAU TETRALOOP, NMR, 3 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*CP*UP*CP*CP*AP*GP*AP*UP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1ATX
 
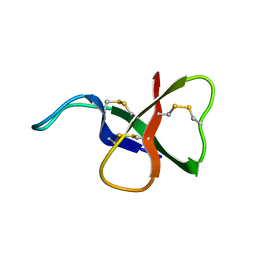 | |
1ATY
 
 | |
1ATZ
 
 | | HUMAN VON WILLEBRAND FACTOR A3 DOMAIN | | Descriptor: | VON WILLEBRAND FACTOR | | Authors: | Huizinga, E.G, Gros, P. | | Deposit date: | 1997-08-15 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the A3 domain of human von Willebrand factor: implications for collagen binding.
Structure, 5, 1997
|
|
1AU0
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC DIACYLAMINOMETHYL KETONE INHIBITOR | | Descriptor: | 1,3-BIS[[N-[(PHENYLMETHOXY)CARBONYL]-L-LEUCYL]AMINO]-2-PROPANONE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1AU1
 
 | | HUMAN INTERFERON-BETA CRYSTAL STRUCTURE | | Descriptor: | INTERFERON-BETA, ZINC ION, alpha-D-quinovopyranose-(1-6)-beta-D-glucopyranose, ... | | Authors: | Karpusas, M, Nolte, M, Lipscomb, W. | | Deposit date: | 1997-09-09 | | Release date: | 1998-06-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human interferon beta at 2.2-A resolution.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AU2
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PROPANONE INHIBITOR | | Descriptor: | 1-[[(4-PHENOXYPHENYL)SULFONYL]AMINO]-3-[[N/N-(4-PYRIDINYLCARBONYL)-L-LEUCYL]AMINO]-2-PROPANOL, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1AU3
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PYRROLIDINONE INHIBITOR | | Descriptor: | 1-[N[(PHENYLMETHOXY)CARBONYL]-L-LEUCYL-4-[[N/N-[(PHENYLMETHOXY)CARBONYL]-/NL-LEUCYL]AMINO]-3-PYRROLIDINONE/N, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally constrained 1,3-diamino ketones: a series of potent inhibitors of the cysteine protease cathepsin K.
J.Med.Chem., 41, 1998
|
|
1AU4
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PYRROLIDINONE INHIBITOR | | Descriptor: | 4-[[N-[(PHENYLMETHOXY)CARBONYL]-/NL/N-LEUCYL]AMINO]-1[(2S)-2-[[[4-(PYRIDINYLMETHOXY)CARBONYL]AMINO]-4-METHYLPENT/NYL]-3-PYRROLIDINONE/N, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally constrained 1,3-diamino ketones: a series of potent inhibitors of the cysteine protease cathepsin K.
J.Med.Chem., 41, 1998
|
|
1AU5
 
 | | SOLUTION STRUCTURE OF INTRASTRAND CISPLATIN-CROSSLINKED DNA OCTAMER D(CCTG*G*TCC):D(GGACCAGG), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*TP*GP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*CP*AP*GP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis.
Biochemistry, 34, 1995
|
|
1AU6
 
 | | SOLUTION STRUCTURE OF DNA D(CATGCATG) INTERSTRAND-CROSSLINKED BY BISPLATIN COMPOUND (1,1/T,T), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIS(TRANS-PLATINUM ETHYLENEDIAMINE DIAMINE CHLORO)COMPLEX, DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Farrell, N, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel DNA structure induced by the anticancer bisplatinum compound crosslinked to a GpC site in DNA.
Nat.Struct.Biol., 2, 1995
|
|
1AU7
 
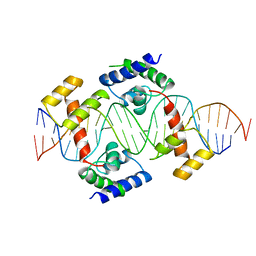 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
1AU8
 
 | | HUMAN CATHEPSIN G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-[(1R)-5-amino-1-phosphonopentyl]-L-prolinamide | | Authors: | Medrano, F.J, Bode, W, Banbula, A, Potempa, J. | | Deposit date: | 1997-09-12 | | Release date: | 1998-10-14 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HUMAN CATHEPSIN G
to be published
|
|
1AU9
 
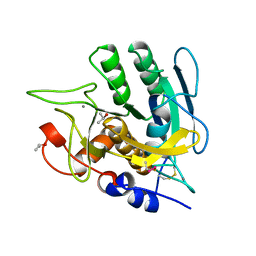 | | SUBTILISIN BPN' MUTANT 8324 IN CITRATE | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN BPN', ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-09-12 | | Release date: | 1997-12-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AUA
 
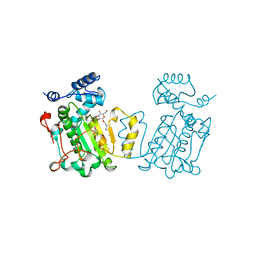 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
