1XII
 
 | |
1XIJ
 
 | |
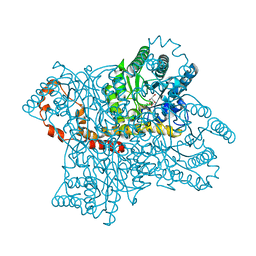
1BPD
 
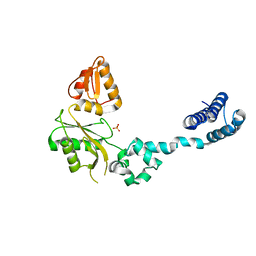 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | 分子名称: | DNA POLYMERASE BETA, PHOSPHATE ION | | 著者 | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | 登録日 | 1994-04-12 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
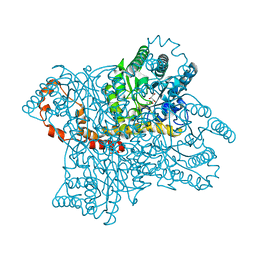
1BPB
 
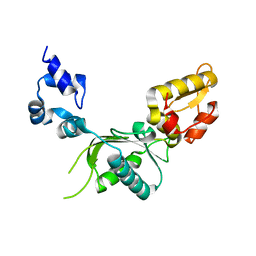 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | 登録日 | 1994-04-12 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1XIE
 
 | |
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | 分子名称: | GLUCOCORTICOID RECEPTOR, ZINC ION | | 著者 | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | 登録日 | 1994-03-15 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
2GDA
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | 分子名称: | GLUCOCORTICOID RECEPTOR, ZINC ION | | 著者 | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | 登録日 | 1994-03-15 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1LST
 
 | |
1NDC
 
 | |
1GLM
 
 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | 分子名称: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | 登録日 | 1994-04-25 | | 公開日 | 1994-06-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
1GLP
 
 | |
1BYB
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | 分子名称: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | 登録日 | 1994-01-25 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1HTG
 
 | |
1BMD
 
 | |
1GPH
 
 | |
2ACU
 
 | | TYROSINE-48 IS THE PROTON DONOR AND HISTIDINE-110 DIRECTS SUBSTRATE STEREOCHEMICAL SELECTIVITY IN THE REDUCTION REACTION OF HUMAN ALDOSE REDUCTASE: ENZYME KINETICS AND THE CRYSTAL STRUCTURE OF THE Y48H MUTANT ENZYME | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bohren, K.M, Grimshaw, C.E, Lai, C.-J, Gabbay, K.H, Petsko, G.A, Harrison, D.H, Ringe, D. | | 登録日 | 1994-04-15 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme.
Biochemistry, 33, 1994
|
|
1CSK
 
 | |
137L
 
 | |
1CRQ
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | 分子名称: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | 登録日 | 1993-11-24 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CHN
 
 | |
1IFM
 
 | |
1IFK
 
 | |
1SMF
 
 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | 分子名称: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | 著者 | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | 登録日 | 1992-10-24 | | 公開日 | 1994-07-31 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
1AKC
 
 | |
216L
 
 | |
